HSC Cell Imaging Core
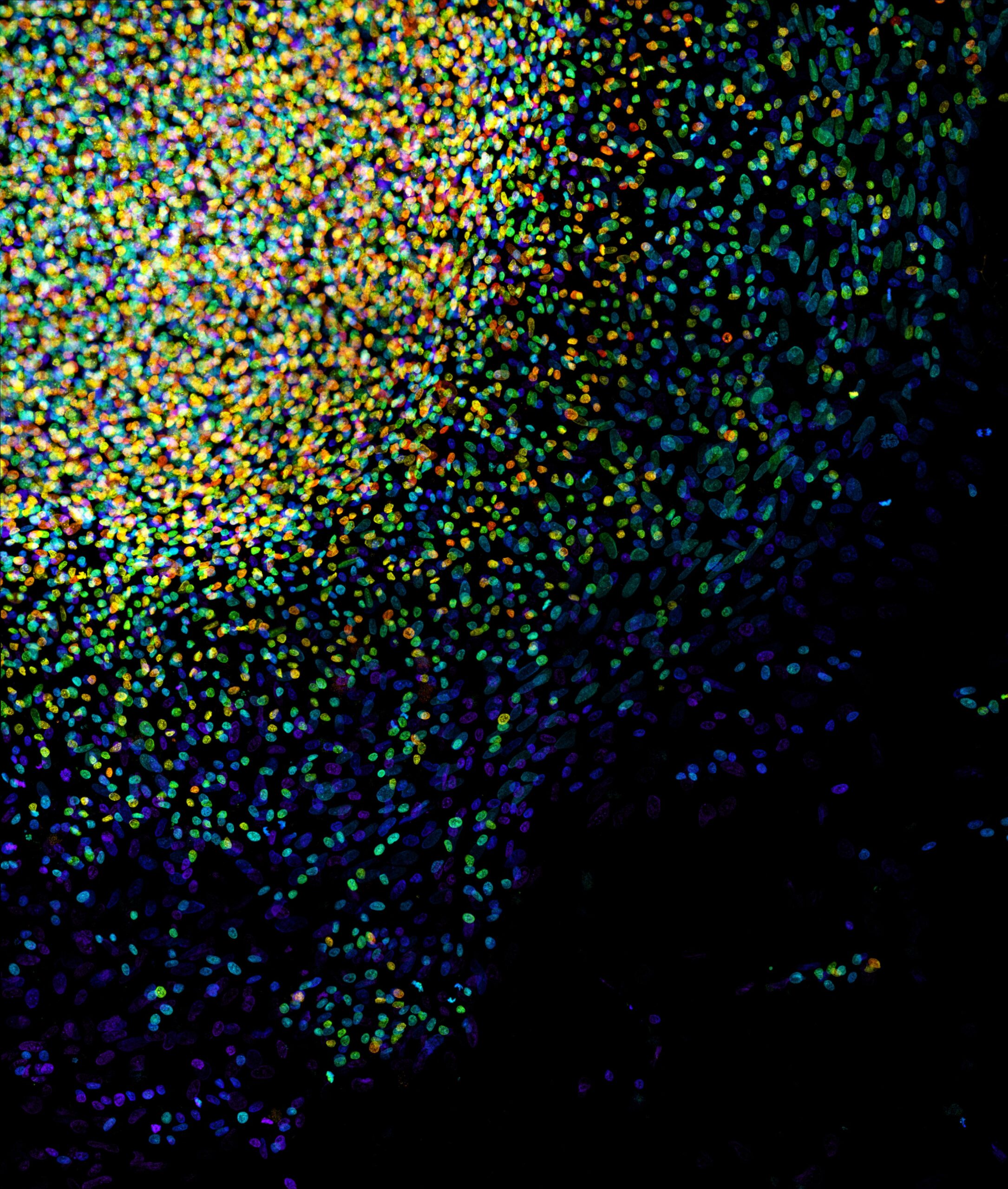
The HSC Cell Imaging Core, with over 20 years of experience, consistently delivers exceptional services to researchers within and beyond the University of Utah, as well as catering to corporate clients seeking access to cutting-edge optical microscopes and advanced imaging analysis software.
As a result of strong and continuous support from both the University and our dedicated users, the HSC Cell Imaging Core has expanded to five major locations across the campus. Each year, more than 300 researchers can readily access our extensive array of over 15 standard and state-of-the-art instruments. We offer comprehensive training for independent instrument use and also provide Assisted Imaging services through our team of experienced core members. In addition, we enthusiastically engage in deep collaborations to assist with complex projects.
We kindly request that patrons acknowledge the HSC Cell Imaging Core in their publications whenever experimental data is generated using our core instruments. This recognition serves to underscore the impact of your work and highlights the vital contributions that the Cell Imaging Core makes. It also plays a crucial role in our efforts to secure grants and funding for acquiring equipment that benefits our user community.
Welcome to Cell Imaging News!
News & Announcements
Instruments Available
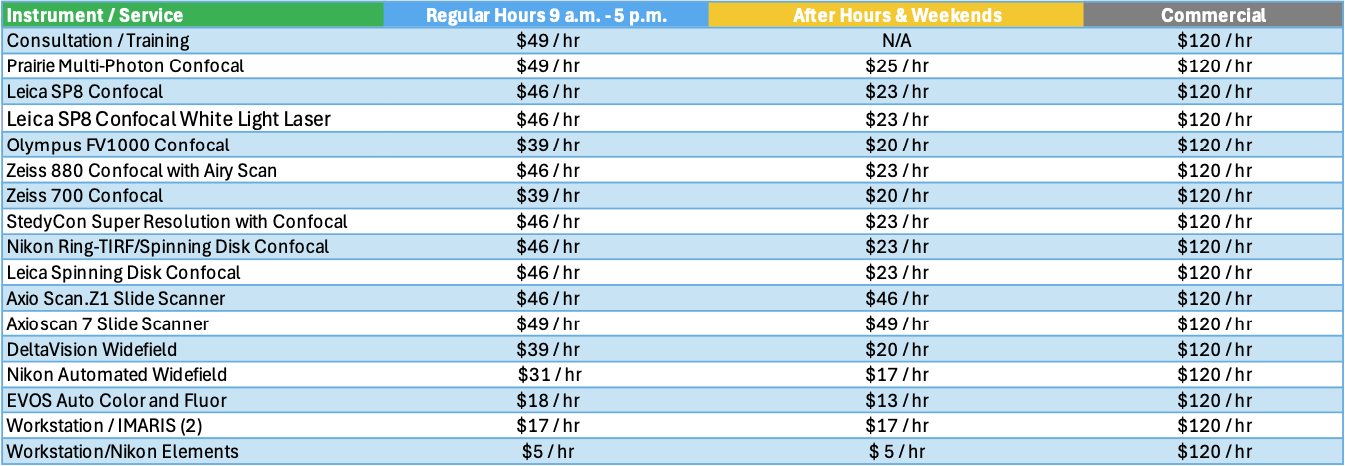
Service Rates
Instruments and workstations can be booked at any time. Bookings can only be cancelled or modified at least 12 hours in advance of the session start time. Booked time has to be used otherwise. To schedule a training or consultation session with a Cell Imaging staff member, a 24-hour lead time is required. Please contact the Cell Imaging staff directly if you need to adjust your scheduling on shorter notice.
Cell Imaging Core Policies Agreement
Requesting Services
Existing users may login directly to the Resource Scheduling System to schedule or order services. This system is cores-wide and uses University of Utah uNID authentication.
Process to Become an HSC Cell Imaging Core User:
- Register as a Core Member: Complete the online Work Authorization Form.
- Complete Laser Safety Training: complete the training module and email the certificate to Cell Imaging Support.
- Sign the Cell Imaging Core Policies Agreement: Read and sign the Cell Imaging Core Policies Agreement and email it to Cell Imaging Support.
- Complete Consultation: Fill out the Patron Consultation Form and schedule a 30-minute consultation with one of the core members via Core Resources Scheduling. A ZOOM meeting will be held.
- Subscribe to the Email List: Join the mailing list to receive important updates and event notifications.
- Book the Instrument and Core Member’s Calendar for initial Training: Simultaneously book the instrument and a core member’s calendar for 2 hours for your first training. Use the Core Resources Scheduling system. (Need help? Instructional video here.)
- Refresher Training: Users who haven’t used the core instruments for over six months or have prior experience must book a 1-hour refresher training.
- Access to the Room: Core members will assist trained users in accessing the room after training.
- Book the Calendar: Schedule time to start using the core instruments.
- 24-Hour Lead Time: A 24-hour lead time is required to book a core member’s calendar.
- Last-Minute Booking: Last-minute bookings are allowed for instrument calendars.
- Cancellation Policy: Schedules cannot be canceled or modified within 12 hours of the experiment’s start time.
- Acknowledge the Core Instrument: If published data is generated using core instruments, acknowledge the Core Instrument. Check the “Publications” tab for details.
Facilities
HSC - Health Science Center - Building 585 - MAP
Offices: 48A, 48E, 55
Labs: 48B, 48D, 54, 56, 60
Phone Number: 801-587-7964
HCI - Huntsman Cancer Institute North - Building 555 - MAP
Offices: 1460
Labs: 1430, 1440, 1470 1480
Phone Number: 801-585-0106
CSC - Crocker Science Center - Building 5 - MAP
Offices: 030
Labs: 032
EEJ - Jones Medical Research - Building 565 - MAP
Labs: 3532, 5100, 5122
ASB - Skaggs Biology Building - Building 82 - MAP
Labs: 230
Hours of Operation
9:00 am to 5:00 pm
Monday - Friday
Contact Email
Cell Imaging Support
Mailing Address
Cell Imaging
Old Skaggs, Bldg 582
30 S 2000 E, Rm 250
Salt Lake City, UT 84112
Citing Our Facility
Acknowledgments
We would like to thank you for acknowledging the our facility. This recognition allows us to highlight the impact of your work and demonstrates the important contributions of our facility makes to research across the University of Utah. The recognition our core receives from your acknowledgments also aids in receiving grants and further funding for equipment and services we can provide to our users.
Self-Run Services / Instrumentation Usage:
In published papers that used instruments at our facility and notably involved staff members please use the following format:
We acknowledge (facility name) at the University of Utah for use of equipment (insert instrument/service details here), and thank (insert any notable staff member – if desired) for their assistance.
Assisted Services:
In published papers where a staff member assisted you in addition to the requested services please use the following format:
We acknowledge (facility name) at the University of Utah for use of equipment (insert instrument/service details here), and thank (insert staff member-required) for their assistance in (service provided).
Collaboration:
For publications resulting from collaborations that assisted with the methodologies, planning process and execution of your experiment in addition to equipment usage we require Co-author attribution on your publication for our facility and any staff members who provided substantial contributions to the originating project.
All Publications
2023
- Anderl, W. J., Pearson, N., Converse, M. I., Yu, S. M., & Monson, K. L. (2023). Strain-induced collagen denaturation is rate dependent in failure of cerebral arteries. Acta Biomaterialia, 164, 282–292. https://doi.org/10.1016/j.actbio.2023.04.032
- Anderson, B., Blair, D., Huff, K., Wisniewski, J., Warner, K. S., & Kauser, K. (2023). Photochemical Modification of the Extracellular Matrix to Alter the Vascular Remodeling Process. Journal of Functional Biomaterials, 14(12), Article 12. https://doi.org/10.3390/jfb14120566
- Carney, K. R., Khan, A. M., Stam, S., Samson, S. C., Mittal, N., Han, S. J., Bidone, T. C., & Mendoza, M. C. (2023a). Nascent adhesions shorten the period of lamellipodium protrusion through the Brownian ratchet mechanism. Molecular Biology of the Cell, mbc.E23-08-0314. https://doi.org/10.1091/mbc.E23-08-0314
- Carney, K. R., Khan, A. M., Stam, S., Samson, S. C., Mittal, N., Han, S. J., Bidone, T. C., & Mendoza, M. C. (2023b). Nascent adhesions shorten the period of lamellipodium protrusion through the Brownian ratchet mechanism. Molecular Biology of the Cell, 34(12), ar115. https://doi.org/10.1091/mbc.E23-08-0314
- Chiola, S., Yang, J., Ullah, H. M. A., Napan, K., Huang, Q., Gamboa, N., Youssef, O., Colman, H., Cheshier, S. H., & Shcheglovitov, A. (2023). Transcriptional changes consistent with altered neuronal differentiation, angiogenesis, and tumor plasticity induced in human subpallial telencephalic organoid-glioblastoma chimeras (p. 2023.05.11.540229). bioRxiv. https://doi.org/10.1101/2023.05.11.540229
- de Hart, N. M. M. P., Petrocelli, J. J., Nicholson, R. J., Yee, E. M., van Onselen, L., Lang, M. J., Bourrant, P.-E., Ferrara, P. J., Bastian, E. D., Ward, L. S., Petersen, B. L., & Drummond, M. J. (2023). Dietary delivery of glycomacropeptide within the whey protein matrix is not effective in mitigating tissue ceramide deposition and obesity in high fat fed mice. Journal of Dairy Science. https://doi.org/10.3168/jds.2023-23914
- de Hart, N. MMP., Petrocelli, J. J., Nicholson, R. J., Yee, E. M., Ferrara, P. J., Bastian, E. D., Ward, L. S., Petersen, B. L., Summers, S. A., & Drummond, M. J. (2023). Palmitate-Induced Inflammation and Myotube Atrophy in C2C12 Cells Are Prevented by the Whey Bioactive Peptide, Glycomacropeptide. The Journal of Nutrition, 153(10), 2915–2928. https://doi.org/10.1016/j.tjnut.2023.08.033
- Digal, L., Samson, S., Stevens, M., Ghorai, A., Kim, H., Mifflin, M., Carney, K., Williamson, D., Um, S., Nagy, G., Oh, D.-C., Mendoza, M., & Roberts, A. (2023). Non-threaded isomers of sungsanpin and ulleungdin lasso peptides inhibit H1299 cancer cell migration [Preprint]. Chemistry. https://doi.org/10.26434/chemrxiv-2023-cfnrh
- Ellis, K. E., Bervoets, S., Smihula, H., Ganguly, I., Vigato, E., Auer, T. O., Benton, R., Litwin-Kumar, A., & Caron, S. J. C. (2023). Evolution of connectivity architecture in the Drosophila mushroom body (p. 2023.02.10.528036). bioRxiv. https://doi.org/10.1101/2023.02.10.528036
- Figueroa, K. P., Gross, C., Atienza, E. B., Paul, S., Gandelman, M., Haack, T., Kakar, N., Sturm, M., Casadei, N., Admard, J., Park, J., Zühlke, C., Hellenbroich, Y., Pozojevic, J., Balachandran, S., Händler, K., Zittel, S., Timmann, D., Erdlenbruch, F., … Pulst, S. M. (2023). GGC expansion in ZFHX3 causes SCA4 and impairs autophagy (p. 2023.10.26.23297560). medRxiv. https://doi.org/10.1101/2023.10.26.23297560
- Guo, L., Jacob, S., Manne, B. K., Kolawole, E. M., Guo, S., Wang, X., Murray, D., Tugolukova, E. A., Portier, I., Kosaka, Y., Barba, C., Rondina, M. T., Evavold, B., Morley, C., Bhatlekar, S., & Bray, P. F. (2023). Actin-bundling protein L-plastin promotes megakaryocyte rigidity and dampens proplatelet formation. Haematologica, 109(1), 331–336. https://doi.org/10.3324/haematol.2023.283016
- Nagarajan N, Capecchi MR. Optogenetic stimulation of mouse Hoxb8 microglia in specific regions of the brain induces anxiety, grooming, or both. Mol Psychiatry. 2023 Apr 10;. doi: 10.1038/s41380-023-02019-w. [Epub ahead of print] PubMed PMID: 37037872.
- Paine, E. L., Skalicky, J. J., Whitby, F. G., Mackay, D. R., Ullman, K. S., Hill, C. P., & Sundquist, W. I. (2023). The Calpain-7 protease functions together with the ESCRT-III protein IST1 within the midbody to regulate the timing and completion of abscission. eLife, 12, e84515. https://doi.org/10.7554/eLife.84515
- Petrocelli, J. J., McKenzie, A. I., de Hart, N. M. M. P., Reidy, P. T., Mahmassani, Z. S., Keeble, A. R., Kaput, K. L., Wahl, M. P., Rondina, M. T., Marcus, R. L., Welt, C. K., Holland, W. L., Funai, K., Fry, C. S., & Drummond, M. J. (2023). Disuse-induced muscle fibrosis, cellular senescence, and senescence-associated secretory phenotype in older adults are alleviated during re-ambulation with metformin pre-treatment. Aging Cell, n/a(n/a), e13936. https://doi.org/10.1111/acel.13936
- Scherer, S. D., Zhao, L., Butterfield, A. J., Yang, C.-H., Cortes-Sanchez, E., Guillen, K. P., Welm, B. E., & Welm, A. L. (2023). Breast cancer PDxO cultures for drug discovery and functional precision oncology. STAR Protocols, 4(3), 102402. https://doi.org/10.1016/j.xpro.2023.102402
- Van Scoyk, A. N., Antelope, O., Franzini, A., Ayer, D. E., Peterson, R. T., Pomicter, A. D., Owen, S. C., & Deininger, M. W. (2023). Bioluminescence Assay of Lysine Deacylase Sirtuin Activity. bioRxiv, 2023.08.10.552871. https://doi.org/10.1101/2023.08.10.552871
2024
- Carey, C. M., Hollins, H. L., Schmid, A. V., & Gagnon, J. A. (2024). Distinct features of the regenerating heart uncovered through comparative single-cell profiling. Biology Open, 13(4), bio060156. https://doi.org/10.1242/bio.060156
- Casey, M. J., Chan, P. P., Li, Q., Jette, C. A., Kohler, M., Myers, B. R., & Stewart, R. A. (2024). A Simple and Scalable Zebrafish Model of Sonic Hedgehog Medulloblastoma (p. 2024.02.03.577834). bioRxiv. https://doi.org/10.1101/2024.02.03.577834
- Collins, B. C., Shapiro, J. B., Scheib, M. M., Musci, R. V., Verma, M., & Kardon, G. (2024). Three-dimensional imaging studies in mice identify cellular dynamics of skeletal muscle regeneration. Developmental Cell. https://doi.org/10.1016/j.devcel.2024.03.017
- de Hart, N. M. M. P., Petrocelli, J. J., Nicholson, R. J., Yee, E. M., van Onselen, L., Lang, M. J., Bourrant, P.-E., Ferrara, P. J., Bastian, E. D., Ward, L. S., Petersen, B. L., & Drummond, M. J. (2024). Dietary delivery of glycomacropeptide within the whey protein matrix is not effective in mitigating tissue ceramide deposition and obesity in mice fed a high-fat diet. Journal of Dairy Science, 107(2), 669–682. https://doi.org/10.3168/jds.2023-23914
- Fennel, Z. J., Bourrant, P.-E., Kurian, A. S., Petrocelli, J. J., de Hart, N. M. M. P., Yee, E. M., Boudina, S., Keirstead, H. S., Nistor, G., Greilach, S. A., Berchtold, N. C., Lane, T. E., & Drummond, M. J. (2024). Stem cell secretome treatment improves whole-body metabolism, reduces adiposity, and promotes skeletal muscle function in aged mice. Aging Cell, n/a(n/a), e14144. https://doi.org/10.1111/acel.14144
- Figueroa, K. P., Gross, C., Buena-Atienza, E., Paul, S., Gandelman, M., Kakar, N., Sturm, M., Casadei, N., Admard, J., Park, J., Zühlke, C., Hellenbroich, Y., Pozojevic, J., Balachandran, S., Händler, K., Zittel, S., Timmann, D., Erdlenbruch, F., Herrmann, L., … Pulst, S. M. (2024). A GGC-repeat expansion in ZFHX3 encoding polyglycine causes spinocerebellar ataxia type 4 and impairs autophagy. Nature Genetics, 1–10. https://doi.org/10.1038/s41588-024-01719-5
- Herstine, J. A., Chang, P.-K., Chornyy, S., Stevenson, T. J., Sunshine, A. C., Nokhrina, K., Rediger, J., Wentz, J., Vetter, T. A., Scholl, E., Holaway, C., Pyne, N. K., Bratasz, A., Yeoh, S., Flanigan, K. M., Bonkowsky, J. L., & Bradbury, A. M. (2024). Evaluation of safety and early efficacy of AAV gene therapy in mouse models of vanishing white matter disease. Molecular Therapy: The Journal of the American Society of Gene Therapy, S1525-0016(24)00212-0. https://doi.org/10.1016/j.ymthe.2024.03.034
- Merrill, C. B., Titos, I., Pabon, M. A., Montgomery, A. B., Rodan, A. R., & Rothenfluh, A. (2024). Iterative assay for transposase-accessible chromatin by sequencing to isolate functionally relevant neuronal subtypes. Science Advances, 10(13), eadi4393. https://doi.org/10.1126/sciadv.adi4393
- Morales, J. C. F., Redfearn, C., Titus, M. A., & Roh-Johnson, M. (2024). Reduced PaxillinB localization to cell-substrate adhesions promotes cell migration in Dictyostelium (p. 2024.03.19.585764). bioRxiv. https://doi.org/10.1101/2024.03.19.585764
- Moriwaki, M., Liu, L., James, E. R., Tolley, N., O’Connora, A. M., Emery, B., Aston, K. I., Campbell, R. A., & Welt, C. K. (2024). Heterozygous Eif4nif1 Stop Gain Mice Replicate the Primary Ovarian Insufficiency Phenotype in Women (p. 2024.04.09.588694). bioRxiv. https://doi.org/10.1101/2024.04.09.588694
- Nwagbo, U., Parvez, S., Maschek, J. A., & Bernstein, P. S. (2024). Elovl4b knockout zebrafish as a model for ocular very-long-chain PUFA deficiency. Journal of Lipid Research, 65(3). https://doi.org/10.1016/j.jlr.2024.100518
- Smith, J. J., Valentino, T. R., Ablicki, A. H., Banerjee, R., Colligan, A. R., Eckert, D. M., Desjardins, G. A., & Diehl, K. L. (2024). A genetically-encoded fluorescent biosensor for visualization of acetyl-CoA in live cells (p. 2023.12.31.573774). bioRxiv. https://doi.org/10.1101/2023.12.31.573774
- Verma, S., Giagnocavo, S. D., Curtin, M. C., Arumugam, M., Osburn-Staker, S. M., Wang, G., Atkinson, A., Nix, D. A., Lum, D. H., Cox, J. E., & Hilgendorf, K. I. (2024). Zinc Alpha-2-Glycoprotein (ZAG/AZGP1) secreted by triple-negative breast cancer promotes tumor microenvironment fibrosis (p. 2024.03.04.583349). bioRxiv. https://doi.org/10.1101/2024.03.04.583349
- Walker, M. F., Zhang, J., Steiner, W., Ku, P.-I., Zhu, J.-F., Michaelson, Z., Yen, Y.-C., Lee, A., Long, A. B., Casey, M. J., Poddar, A., Nelson, I. B., Arveseth, C. D., Nagel, F., Clough, R., LaPotin, S., Kwan, K. M., Schulz, S., Stewart, R. A., … Myers, B. R. (2024). GRK2 Kinases in the Primary Cilium Initiate SMOOTHENED-PKA Signaling in the Hedgehog Cascade (p. 2023.05.10.540226). bioRxiv. https://doi.org/10.1101/2023.05.10.540226
- Wallis, G. J., Bell, L. A., Wagner, J. N., Buxton, L., Balachandar, L., & Wilcox, K. S. (2024). Reactive microglia fail to respond to environmental damage signals in a viral-induced mouse model of temporal lobe epilepsy (p. 2024.03.06.583768). bioRxiv. https://doi.org/10.1101/2024.03.06.583768
2022
- Hagen-Lillevik, S., Johnson, J., & Lai, K. (2022). Early postnatal alterations in follicular stress response and survival in a mouse model of Classic Galactosemia. Journal of Ovarian Research, 15(1), 122. https://doi.org/10.1186/s13048-022-01049-2
- Hagen-Lillevik, S., Johnson, J., Siddiqi, A., Persinger, J., Hale, G., & Lai, K. (2022). Harnessing the Power of Purple Sweet Potato Color and Myo-Inositol to Treat Classic Galactosemia. International Journal of Molecular Sciences, 23(15), 8654. https://doi.org/10.3390/ijms23158654
- Hoffman, L. M., Jensen, C. C., & Beckerle, M. C. (2022). Phosphorylation of the small heat shock protein HspB1 regulates cytoskeletal recruitment and cell motility. Molecular Biology of the Cell, 33(11), ar100. https://doi.org/10.1091/mbc.E22-02-0057
- LaJoie, D., Turkmen, A. M., Mackay, D. R., Jensen, C. C., Aksenova, V., Niwa, M., Dasso, M., & Ullman, K. S. (2022). A role for Nup153 in nuclear assembly reveals differential requirements for targeting of nuclear envelope constituents. Molecular Biology of the Cell, 33(13), ar117. https://doi.org/10.1091/mbc.E22-05-0189
- Rush, C. M., Blanchard, Z., Polaski, J. T., Osborne, K. S., Osby, K., Vahrenkamp, J. M., Yang, C.-H., Lum, D. H., Hagan, C. R., Leslie, K. K., Pufall, M. A., Thiel, K. W., & Gertz, J. (2022). Characterization of HCI-EC-23 a novel estrogen- and progesterone-responsive endometrial cancer cell line. Scientific Reports, 12(1), Article 1. https://doi.org/10.1038/s41598-022-24211-8
- Sefton, E. M., Gallardo, M., Tobin, C. E., Collins, B. C., Colasanto, M. P., Merrell, A. J., & Kardon, G. (2022). Fibroblast-derived Hgf controls recruitment and expansion of muscle during morphogenesis of the mammalian diaphragm. ELife, 11, e74592. https://doi.org/10.7554/eLife.74592
- Simeone, C. A., Wilkerson, J. L., Poss, A. M., Banks, J. A., Varre, J. V., Guevara, J. L., Hernandez, E. J., Gorsi, B., Atkinson, D. L., Turapov, T., Frodsham, S. G., Morales, J. C. F., O’Neil, K., Moore, B., Yandell, M., Summers, S. A., Krolewski, A. S., Holland, W. L., & Pezzolesi, M. G. (2022). A dominant negative ADIPOQ mutation in a diabetic family with renal disease, hypoadiponectinemia, and hyperceramidemia. Npj Genomic Medicine, 7(1), Article 1. https://doi.org/10.1038/s41525-022-00314-z
- Smith, M. A., Blankman, E., Jensen, C. C., Hoffman, L. M., Ullman, K. S., & Beckerle, M. C. (2022). Nuclear pore complexes concentrate on Actin/LINC/Lamin nuclear lines in response to mechanical stress in a SUN1 dependent manner. Heliyon, 8(12), e12147. https://doi.org/10.1016/j.heliyon.2022.e12147
- Wang, Y., Chiola, S., Yang, G., Russell, C., Armstrong, C. J., Wu, Y., Spampanato, J., Tarboton, P., Ullah, H. M. A., Edgar, N. U., Chang, A. N., Harmin, D. A., Bocchi, V. D., Vezzoli, E., Besusso, D., Cui, J., Cattaneo, E., Kubanek, J., & Shcheglovitov, A. (2022). Modeling human telencephalic development and autism-associated SHANK3 deficiency using organoids generated from single neural rosettes. Nature Communications, 13(1), Article 1. https://doi.org/10.1038/s41467-022-33364-z
- Wenzel, D. M., Mackay, D. R., Skalicky, J. J., Paine, E. L., Miller, M. S., Ullman, K. S., & Sundquist, W. I. (2022). Comprehensive analysis of the human ESCRT-III-MIT domain interactome reveals new cofactors for cytokinetic abscission. ELife, 11, e77779. https://doi.org/10.7554/eLife.77779
- Zhang, C., Jin, Y., Marchetti, M., Lewis, M. R., Hammouda, O. T., & Edgar, B. A. (2022). EGFR signaling activates intestinal stem cells by promoting mitochondrial biogenesis and β-oxidation. Current Biology. https://doi.org/10.1016/j.cub.2022.07.003
2023
- Almanzar, D. E., Gordon, S. G., Bristow, C., Hamrick, A., von Diezmann, L., Liu, H., & Rog, O. (2023). Meiotic DNA exchanges in C. elegans are promoted by proximity to the synaptonemal complex. Life Science Alliance, 6(4), e202301906. https://doi.org/10.26508/lsa.202301906
- Anderl, W. J., Pearson, N., Converse, M. I., Yu, S. M., & Monson, K. L. (2023). Strain-induced collagen denaturation is rate dependent in failure of cerebral arteries. Acta Biomaterialia. https://doi.org/10.1016/j.actbio.2023.04.032
- Ellis, K. E., Smihula, H., Ganguly, I., Vigato, E., Bervoets, S., Auer, T. O., Benton, R., Litwin-Kumar, A., & Caron, S. J. C. (2023). Evolution of connectivity architecture in the Drosophila mushroom body [Preprint]. Neuroscience. https://doi.org/10.1101/2023.02.10.528036
- Eshima, H., Shahtout, J. L., Siripoksup, P., Pearson, M. J., Mahmassani, Z. S., Ferrara, P. J., Lyons, A. W., Maschek, J. A., Peterlin, A. D., Verkerke, A. R. P., Johnson, J. M., Salcedo, A., Petrocelli, J. J., Miranda, E. R., Anderson, E. J., Boudina, S., Ran, Q., Cox, J. E., Drummond, M. J., & Funai, K. (2023). Lipid hydroperoxides promote sarcopenia through carbonyl stress. ELife, 12, e85289. https://doi.org/10.7554/eLife.85289
- Espino-Sanchez, T. J., Wienkers, H., Marvin, R. G., Nalder, S.-A., García-Guerrero, A. E., VanNatta, P. E., Jami-Alahmadi, Y., Mixon Blackwell, A., Whitby, F. G., Wohlschlegel, J. A., Kieber-Emmons, M. T., Hill, C. P., & Sigala, P. A. (2023). Direct tests of cytochrome c and c1 functions in the electron transport chain of malaria parasites. Proceedings of the National Academy of Sciences of the United States of America, 120(19), e2301047120. https://doi.org/10.1073/pnas.2301047120
- Ferrara, P. J., Reidy, P. T., Petrocelli, J. J., Yee, E. M., Fix, D. K., Mahmassani, Z. S., Montgomery, J. A., McKenzie, A. I., de Hart, N. M. M. P., & Drummond, M. J. (2023). Global deletion of CCL2 has adverse impacts on recovery of skeletal muscle fiber size and function and is muscle-specific. Journal of Applied Physiology. https://doi.org/10.1152/japplphysiol.00444.2022
- He, Y., Anderson, B., Hu, Q., Hayes, R. B., Huff, K., Isaacson, J., Warner, K. S., Hauser, H., Greenberg, M., Chandra, V., Kauser, K., & Berceli, S. A. (2023). Photochemically Aided Arteriovenous Fistula Creation to Accelerate Fistula Maturation. International Journal of Molecular Sciences, 24(8), Article 8. https://doi.org/10.3390/ijms24087571
- Kidwell, C. U., Casalini, J. R., Pradeep, S., Scherer, S. D., Greiner, D., Bayik, D., Watson, D. C., Olson, G. S., Lathia, J. D., Johnson, J. S., Rutter, J., Welm, A. L., Zangle, T. A., & Roh-Johnson, M. (2023). Transferred mitochondria accumulate reactive oxygen species, promoting proliferation. ELife, 12, e85494. https://doi.org/10.7554/eLife.85494
- Merrill, C. B., Titos, I., Pabon, M. A., Montgomery, A. B., Rodan, A. R., & Rothenfluh, A. (2023). Iterative assay for transposase-accessible chromatin by sequencing to isolate functionally relevant neuronal subtypes (p. 2023.04.14.536950). bioRxiv. https://doi.org/10.1101/2023.04.14.536950
- Petrocelli, J. J., Hart, N. M. M. P. de, Lang, M. J., Yee, E. M., Ferrara, P. J., Fix, D. K., Chaix, A., Funai, K., & Drummond, M. J. (2023). Cellular senescence and disrupted proteostasis induced by myotube atrophy are prevented with low-dose metformin and leucine cocktail. Aging, 15. https://doi.org/10.18632/aging.204600
- Preston, A. J., Rogers, A., Sharp, M., Mitchell, G., Toruno, C., Barney, B. B., Donovan, L. N., Bly, J., Kennington, R., Payne, E., Iovino, A., Furukawa, G., Robinson, R., Shamloo, B., Buccilli, M., Anders, R., Eckstein, S., Fedak, E. A., Wright, T., … Abegglen, L. M. (2023). Elephant TP53-RETROGENE 9 induces transcription-independent apoptosis at the mitochondria. Cell Death Discovery, 9(1), Article 1. https://doi.org/10.1038/s41420-023-01348-7
- Rice, M. C., Little, J. H., Forrister, D. L., Machado, J., Clark, N. L., & Gagnon, J. A. (2023). Gadusol is a maternally provided sunscreen that protects fish embryos from DNA damage [Preprint]. Developmental Biology. https://doi.org/10.1101/2023.01.30.526370
- Stover, J. D., Trone, M. A., Lawrence, B., & Bowles, R. D. (2023). Multiplex epigenome editing of ion channel expression in nociceptive neurons abolished degenerative IVD-conditioned media-induced mechanical sensitivity. JOR SPINE, n/a(n/a), e1253. https://doi.org/10.1002/jsp2.1253
- Su, G., Farhat, R., Laxman, A. K., Chapman-Natewa, K., Nelson, I. E., & Chan, O. (2023). Astrocyte glycogen is a major source of hypothalamic lactate in rats with recurrent hypoglycemia. Diabetes, db220902. https://doi.org/10.2337/db22-0902
- Warde, K. M., Smith, L. J., Liu, L., Stubben, C. J., Lohman, B. K., Willett, P. W., Ammer, J. L., Castaneda-Hernandez, G., Imodoye, S. O., Zhang, C., Jones, K. D., Converso-Baran, K., Ekiz, H. A., Barry, M., Clay, M. R., Kiseljak-Vassiliades, K., Giordano, T. J., Hammer, G. D., & Basham, K. J. (2023). Senescence-induced immune remodeling facilitates metastatic adrenal cancer in a sex-dimorphic manner. Nature Aging, 1–20. https://doi.org/10.1038/s43587-023-00420-2
- Xue, Q., Varady, S. R. S., Waddell, T. Q. A., Roman, M. R., Carrington, J., & Roh-Johnson, M. (2023). Lack of Paxillin phosphorylation promotes single-cell migration in vivo. The Journal of Cell Biology, 222(3), e202206078. https://doi.org/10.1083/jcb.202206078
2021
- Fadul, J., Zulueta-Coarasa, T., Slattum, G. M., Redd, N. M., Jin, M. F., Redd, M. J., Daetwyler, S., Hedeen, D., Huisken, J., & Rosenblatt, J. (2021). KRas-transformed epithelia cells invade and partially dedifferentiate by basal cell extrusion. Nat Commun, 12(1), 7180. doi:10.1038/s41467-021-27513-z
- Falekun, S., Sepulveda, J., Jami-Alahmadi, Y., Park, H., Wohlschlegel, J. A., & Sigala, P. A. (2021). Divergent acyl carrier protein decouples mitochondrial Fe-S cluster biogenesis from fatty acid synthesis in malaria parasites. Elife, 10. doi:10.7554/eLife.71636
- Ferrara, P. J., Verkerke, A. R. P., Maschek, J. A., Shahtout, J. L., Siripoksup, P., Eshima, H., Johnson, J. M., Petrocelli, J. J., Mahmassani, Z. S., Green, T. D., McClung, J. M., Cox, J. E., Drummond, M. J., & Funai, K. (2021). Low lysophosphatidylcholine induces skeletal muscle myopathy that is aggravated by high-fat diet feeding. Faseb Journal, 35(10). doi:10.1096/fj.202101104R
- Fix, D. K., Ekiz, H. A., Petrocelli, J. J., Mckenzie, A. M., Mahmassani, Z. S., O'Connell, R. M., & Drummond, M. J. (2021). Disrupted macrophage metabolic reprogramming in aged soleus muscle during early recovery following disuse atrophy. Aging Cell, 20(9). doi:10.1111/acel.13448
- Fix, D. K., Mahmassani, Z. S., Petrocelli, J. J., de Hart, N. M. M. P., Ferrara, P. J., Painter, J. S., Nistor, G., Lane, T. E., Keirstead, H. S., & Drummond, M. J. (2021). Reversal of deficits in aged skeletal muscle during disuse and recovery in response to treatment with a secrotome product derived from partially differentiated human pluripotent stem cells. Geroscience, 43(6), 2635-2652. doi:10.1007/s11357-021-00423-0
- Happ, J. T., Arveseth, C. D., Bruystens, J., Bertinetti, D., Nelson, I. B., Olivieri, C., Hedeen, D. S., Zhu, J.-F., Capener, J. L., Bröckel, J. W., Vu, L., King, C. C., Ruiz-Perez, V. L., Veglia, G., Herberg, F. W., Taylor, S. S., & Myers, B. R. (2021). A PKA Inhibitor Motif within Smoothened Controls Hedgehog Signal Transduction. bioRxiv. doi:10.1101/2021.07.05.451193
- Kidwell, C. U., Casalini, J. R., Pradeep, S., Scherer, S. D., Greiner, D., Johnson, J. S., Olson, G. S., Rutter, J., Welm, A. L., Zangle, T. A., & Roh-Johnson, M. (2021). Laterally transferred macrophage mitochondria act as a signaling source promoting cancer cell proliferation. bioRxiv. doi:10.1101/2021.08.10.455713
- Kursel, L. E., Cope, H. D., & Rog, O. (2021). Unconventional conservation reveals structure-function relationships in the synaptonemal complex. Elife, 10. doi:10.7554/eLife.72061
- Lai, S. A., Gundlapalli, H., Ekiz, H. A., Jiang, A., Fernandez, E., & Welm, A. L. (2021). Blocking Short-Form Ron Eliminates Breast Cancer Metastases through Accumulation of Stem-Like CD4(+) T Cells That Subvert Immunosuppression. Cancer Discovery, 11(12), 3178-3197. doi:10.1158/2159-8290.CD-20-1172
- Liu, H., Gordon, S. G., & Rog, O. (2021). Heterologous synapsis in C. elegans is regulated by meiotic double-strand breaks and crossovers. Chromosoma, 130(4), 237-250. doi:10.1007/s00412-021-00763-y
- Marchetti, M., Zhang, C., & Edgar, B. A. (2021). An improved organ explant culture method reveals stem cell lineage dynamics in the adult Drosophila intestine. bioRxiv. doi:10.1101/2021.12.17.473114
- Mathew, B., Aoyagi, K. L., & Fisher, M. A. (2021). Yersinia pestis Lipopolysaccharide Remodeling Confers Resistance to a Xenopsylla cheopis Cecropin. Acs Infectious Diseases, 7(8), 2536-2545. doi:10.1021/acsinfecdis.1c00275
- Nicholson, R. J., Poss, A. M., Maschek, J. A., Cox, J. E., Hopkins, P. N., Hunt, S. C., Playdon, M. C., Holland, W. L., & Summers, S. A. (2021). Characterizing a Common CERS2 Polymorphism in a Mouse Model of Metabolic Disease and in Subjects from the Utah CAD Study. Journal of Clinical Endocrinology & Metabolism, 106(8), E3098-E3109. doi:10.1210/clinem/dgab155
- Ost, K. S., O'Meara, T. R., Stephens, W. Z., Chiaro, T., Zhou, H., Penman, J., Bell, R., Catanzaro, J. R., Song, D., Singh, S., Call, D. H., Hwang-Wong, E., Hanson, K. E., Valentine, J. F., Christensen, K. A., O'Connell, R. M., Cormack, B., Ibrahim, A. S., Palm, N. W., Noble, S. M., & Round, J. L. (2021). Adaptive immunity induces mutualism between commensal eukaryotes. Nature, 596(7870), 114-118. doi:10.1038/s41586-021-03722-w
- Petrocelli, J. J., Mahmassani, Z. S., Fix, D. K., Montgomery, J. A., Reidy, P. T., McKenzie, A. I., de Hart, N. M, Ferrara, P. J., Kelley, J. J., Eshima, H., Funai, K., & Drummond, M. J. (2021). Metformin and leucine increase satellite cells and collagen remodeling during disuse and recovery in aged muscle. Faseb Journal, 35(9). doi:10.1096/fj.202100883R
- Rheinemann, L., Downhour, D. M., Bredbenner, K., Mercenne, G., Davenport, K. A., Schmitt, P. T., Necessary, C. R., McCullough, J., Schmitt, A. P., Simon, S. M., Sundquist, W. I., & Elde, N. C. (2021). RetroCHMP3 blocks budding of enveloped viruses without blocking cytokinesis. Cell, 184(21), 5419-5431.e16. doi:10.1016/j.cell.2021.09.008
- Tamamouna, V., Rahman, M. M., Petersson, M., Charalambous, I., Kux, K., Mainor, H., Bolender, V., Isbilir, B., Edgar, B. A., & Pitsouli, C. (2021). Remodelling of oxygen-transporting tracheoles drives intestinal regeneration and tumorigenesis in Drosophila. Nat Cell Biol, 23(5), 497-510. doi:10.1038/s41556-021-00674-1
- Zhang, P., Katzaroff, A. J., Buttitta, L. A., Ma, Y., Jiang, H., Nickerson, D. W., Øvrebø, J. I., & Edgar, B. A. (2021). The Krüppel-like factor Cabut has cell cycle regulatory properties similar to E2F1. Proc Natl Acad Sci U S A, 118(7). doi:10.1073/pnas.2015675118
2022
- Batot, G., Metcalf, C., Bell, L., Pauletti, A., Wilcox, K., & Bröer, S. (2022). A Model for Epilepsy of Infectious Etiology using Theiler's Murine Encephalomyelitis Virus. Journal of Visualized Experiments. doi:10.3791/63673
- Cho, J. M., Park, S., Kwon, O. S., La Salle, D. T., Cerbie, J., Fermoyle, C. C., Morgan, D., Nelson, A., Bledsoe, A., Bharath, L. P., Tandar, M., Kunapuli, S. P., Richardson, R. S., Anandh Babu, P. V., Mookherjee, S., Kishore, B. K., Wang, F., Yang, T., Boudina, S., Trinity, J. D., & Symons, J. D. (2022). Activating P2Y1 receptors improves function in arteries with repressed autophagy. Cardiovascular Research. doi:10.1093/cvr/cvac061
- Figueroa, K., Anderson, C. J., Paul, S., Dansithong, W., Gandelman, M., Scoles, D. R., & Pulst, S. M. (2022). Slc9a6 mutation causes Purkinje cell loss and ataxia in the shaker rat. bioRxiv. doi:10.1101/2022.03.28.486143
- Guillen, K. P., Fujita, M., Butterfield, A. J., Scherer, S. D., Bailey, M. H., Chu, Z., DeRose, Y. S., Zhao, L., Cortes-Sanchez, E., Yang, C. H., Toner, J., Wang, G., Qiao, Y., Huang, X., Greenland, J. A., Vahrenkamp, J. M., Lum, D. H., Factor, R. E., Nelson, E. W., Matsen, C. B., Poretta, J. M., Rosenthal, R., Beck, A. C., Buys, S. S., Vaklavas, C., Ward, J. H., Jensen, R. L., Jones, K. B., Li, Z., Oesterreich, S., Dobrolecki, L. E., Pathi, S. S., Woo, X. Y., Berrett, K. C., Wadsworth, M. E., Chuang, J. H., Lewis, M. T., Marth, G. T., Gertz, J., Varley, K. E., Welm, B. E., & Welm, A. L. (2022). A human breast cancer-derived xenograft and organoid platform for drug discovery and precision oncology. Nat Cancer, 3(2), 232-250. doi:10.1038/s43018-022-00337-6
- Hill, J. H., Massaquoi, M. S., Sweeney, E. G., Wall, E. S., Jahl, P., Bell, R., Kallio, K., Derrick, D., Murtaugh, L. C., Parthasarathy, R., Remington, S. J., Round, J. L., & Guillemin, K. (2022). A microbiota membrane disrupter disseminates to the pancreas and increases β-cell mass. bioRxiv. doi:10.1101/2022.03.24.485696
- Ingram, K., Samson, S. C., Zewdu, R., Zitnay, R. G., Snyder, E. L., & Mendoza, M. C. (2022). NKX2-1 controls lung cancer progression by inducing DUSP6 to dampen ERK activity. Oncogene, 41(2), 293-300. doi:10.1038/s41388-021-02076-x
- Jensen, O., Trivedi, S., Meier, J. D., Fairfax, K. C., Hale, J. S., & Leung, D. T. (2022). A subset of follicular helper-like MAIT cells can provide B cell help and support antibody production in the mucosa. Science Immunology, 7(67). doi:10.1126/sciimmunol.abe8931
- Kaur, K., Mohammadpour, R., Ghandehari, H., Reilly, C. A., Paine, R., & Kelly, K. E. (2022). Effect of combustion particle morphology on biological responses in a Co-culture of human lung and macrophage cells. Atmospheric Environment, 284, 119194. doi:10.1016/j.atmosenv.2022.119194
- Kaur, K., Mohammadpour, R., Sturrock, A., Ghandehari, H., Reilly, C., Paine, R., & Kelly, K. E. (2022). Comparison of biological responses between submerged, pseudo-air-liquid interface, and air-liquid interface exposure of A549 and differentiated THP-1 co-cultures to combustion-derived particles. Journal of Environmental Science and Health, Part A, 1-12. doi:10.1080/10934529.2022.2083429
- LaBelle, S. A., Dinkins, S. S., Hoying, J. B., Budko, E. V., Rauff, A., Strobel, H. A., Lin, A. H., & Weiss, J. A. (2022). Matrix anisotropy promotes angiogenesis in a density-dependent manner. Am J Physiol Heart Circ Physiol, 322(5), H806-H818. doi:10.1152/ajpheart.00072.2022
- Lam, G., Beebe, K., & Thummel, C. S. (2022). A direct-drive GFP reporter for studies of tracheal development in. Fly (Austin), 16(1), 105-110. doi:10.1080/19336934.2022.2030191
- Lin, Y., Perovanovic, J., Kong, Y., Igyarto, B. Z., Zurawski, S., Tantin, D., Zurawski, G., Bettini, M., & Bettini, M. L. (2022). Antibody-Mediated Targeting of a Hybrid-Insulin-Peptide Towards Neonatal Thymic Langerin+ Cells Enhances T Cell Central Tolerance and Delays Autoimmune Diabetes. Diabetes. doi:10.2337/db21-1069
- McKenzie, A. I., Mahmassani, Z. S., Petrocelli, J. J., de Hart, N. M. M. P., Fix, D. K., Ferrara, P. J., LaStayo, P. C., Marcus, R. L., Rondina, M. T., Summers, S. A., Johnson, J. M., Trinity, J. D., Funai, K., & Drummond, M. J. (2022). Short-term exposure to a clinical dose of metformin increases skeletal muscle mitochondrial H2O2 emission and production in healthy, older adults: A randomized controlled trial. Experimental gerontology, 163, 111804. doi:10.1016/j.exger.2022.111804
- Meng, F., Fleming, B. A., Jia, X., Rousek, A. A., Mulvey, M. A., & Ward, D. M. (2022). Lysosomal iron recycling in mouse macrophages is dependent upon both LcytB and Steap3 reductases. Blood Adv, 6(6), 1692-1707. doi:10.1182/bloodadvances.2021005609
- Nie, X., Munyoki, S. K., Sukhwani, M., Schmid, N., Missel, A., Emery, B. R., DonorConnect, Stukenborg, J., Mayerhofer, A., Orwig, K. E., Aston, K. I., Hotaling, J. M., Cairns, B. R., & Guo, J. (2022). Single-cell analysis of human testis aging and correlation with elevated body mass index. Developmental Cell, 57(9), 1160-1176.e5. doi:10.1016/j.devcel.2022.04.004
- Okada, M., Rajaram, K., Swift, R. P., Mixon, A., Maschek, J. A., Prigge, S. T., & Sigala, P. A. (2022). Critical role for isoprenoids in apicoplast biogenesis by malaria parasites. Elife, 11. doi:10.7554/eLife.73208
- Øvrebø, J. I., Bradley-Gill, M., Zielke, N., Kim, M., Marchetti, M., Bohlen, J., Lewis, M., van Straaten, M., Moon, N., & Edgar, B. A. (2022). Translational control of E2f1 regulates the Drosophila cell cycle. Proc Natl Acad Sci U S A, 119(4). doi:10.1073/pnas.2113704119
- Van Deren, D. A., De, S., Xu, B., Eschenbacher, K. M., Zhang, S., & Capecchi, M. R. (2022). Defining the Hoxb8 cell lineage during murine definitive hematopoiesis. Development, 149(8). doi:10.1242/dev.200200
- Wenzel, D. M., Mackay, D. R., Skalicky, J. J., Paine, E. L., Miller, M. S., Ullman, K. S., & Sundquist, W. I. (2022). Comprehensive analysis of the human ESCRT-III-MIT domain interactome reveals new cofactors for cytokinetic abscission. bioRxiv. doi:10.1101/2022.02.09.477148
- Wojtalewicz, S., Vizmeg, J., Erickson, S., Lade, C., Shea, J., Sant, H., Magda, J., Gale, B., Agarwal, J., & Davis, B. (2022). Evaluating the influence of particle morphology and density on the viscosity and injectability of a novel long-acting local anesthetic suspension. J Biomater Appl. doi:10.1177/08853282221106486
- Xue, Q., Varady, S. R. S., Waddell, T. Q. A., Carrington, J., & Roh-Johnson, M. (2022). Focal adhesion-based cell migration is differentially regulated in vivo versus in vitro by Paxillin phosphorylation. bioRxiv. doi:10.1101/2022.03.02.482703
- Yang, G., Parker, E., Gorsi, B., Liebowitz, M., Maguire, C., King, J. B., Coon, H., Lopez-Larson, M., Anderson, J., Yandell, M., & Shcheglovitov, A. (2022). Neurite outgrowth deficits caused by rare PLXNB1 mutation in pediatric bipolar disorder. medRxiv. doi:10.1101/2022.05.06.22274499
- Zhao, H., Pomicter, A. D., Eiring, A. M., Franzini, A., Ahmann, J., Hwang, J.-Y., Senina, A., Helton, B., Iyer, S., Yan, D., Khorashad, J. S., Zabriskie, M. S., Agarwal, A., Redwine, H. M., Bowler, A. D., Clair, P. M., McWeeney, S. K., Druker, B. J., Tyner, J. W., Stirewalt, D. L., Oehler, V. G., Varambally, S., Berrett, K. C., Vahrenkamp, J. M., Gertz, J., Varley, K. E., Radich, J. P., & Deininger, M. W. (2022). MS4A3 promotes differentiation in chronic myeloid leukemia by enhancing common β-chain cytokine receptor endocytosis. Blood, 139(5), 761–778. doi:10.1182/blood.2021011802
2020
- Ahmed, S., Maldera, J., Krunic, D., Paiva-Silva, G., Penalva, C., Teleman, A., & Edgar, B. (2020). Fitness trade-offs incurred by ovary-to-gut steroid signalling in Drosophila. Nature, 584(7821), 415-+. doi:10.1038/s41586-020-2462-y
- Auer, T. O., Khallaf, M. A., Silbering, A. F., Zappia, G., Ellis, K., Álvarez-Ocaña, R., Arguello, J. R., Hansson, B. S., Jefferis, G. S. X.E, Caron, S. J. C., Knaden, M., & Benton, R. (2020). Olfactory receptor and circuit evolution promote host specialization. Nature, 579(7799), 402-408. doi:10.1038/s41586-020-2073-7
- Bell, L. A., Wallis, G. J., & Wilcox, K. S. (2020). Reactivity and increased proliferation of NG2 cells following central nervous system infection with Theiler's murine encephalomyelitis virus. J Neuroinflammation, 17(1), 369. doi:10.1186/s12974-020-02043-5
- Bhatlekar, S., Manne, B. K., Basak, I., Edelstein, L. C., Tugolukova, E., Stoller, M. L., Cody, M. J., Morley, S. C., Nagalla, S., Weyrich, A. S., Rowley, J. W., O'Connell, R. M.,Rondina, M. T., Campbell, R. A., & Bray, P. F. (2020). miR-125a-5p regulates megakaryocyte proplatelet formation via the actin-bundling protein L-plastin. Blood, 136(15), 1760-1772. doi:10.1182/blood.2020005230
- Davis, B., Erickson, S., Wojtalewicz, S., Simpson, A., Metcalf, C., Sant, H., Shea, J., Gale, B., & Agarwal, J. (2020). Entrapping bupivacaine-loaded emulsions in a crosslinked-hydrogel increases anesthetic effect and duration in a rat sciatic nerve block model. International Journal of Pharmaceutics, 588. doi:10.1016/j.ijpharm.2020.119703
- Dong, Z. M., Lin, E., Wechsler, M. E., Weller, P. F., Klion, A. D., Bochner, B. S., Delker, D. A., Hazel, M. W., Fairfax, K., Khoury, P., Akuthota, P., Merkel, P. A., Dyer, A. M., Langford, C., Specks, U., Gleich, G. J., Chinchilli, V. M., Raby, B., Yandell, M., & Clayton, F. (2020). Pulmonary Eosinophilic Granulomatosis with Polyangiitis Has IgG4 Plasma Cells and Immunoregulatory Features. Am J Pathol, 190(7), 1438-1448. doi:10.1016/j.ajpath.2020.03.005
- Eshima, H., Siripoksup, P., Mahmassani, Z., Johnson, J., Ferrara, P., Verkerke, A., Salcedo, A., Drummond, M.J., & Funai, K. (2020). Neutralizing mitochondrial ROS does not rescue muscle atrophy induced by hindlimb unloading in female mice. Journal of Applied Physiology, 129(1), 124-132. doi:10.1152/japplphysiol.00456.2019
- Hoffman, L. M., Smith, M. A., Jensen, C. C., Yoshigi, M., Blankman, E., Ullman, K. S., & Beckerle, M. C. (2020). Mechanical stress triggers nuclear remodeling and the formation of transmembrane actin nuclear lines with associated nuclear pore complexes. Mol Biol Cell, 31(16), 1774-1787. doi:10.1091/mbc.E19-01-0027
- Keefe, M. D., Soderholm, H. E., Shih, H. Y., Stevenson, T. J., Glaittli, K. A., Bowles, D. M., Scholl, E., Colby, S., Merchant, S., Hsu, E. W., & Bonkowsky, J. L. (2020). Vanishing white matter disease expression of truncated EIF2B5 activates induced stress response. Elife, 9. doi:10.7554/eLife.56319
- Li, J., Mahoney, B. D., Jacob, M. S., & Caron, S. J. C. (2020). Visual Input into the Drosophila melanogaster Mushroom Body. Cell Rep, 32(11), 108138. doi:10.1016/j.celrep.2020.108138
- Okada, M., Guo, P., Nalder, S. A., & Sigala, P. A. (2020). Doxycycline has distinct apicoplast-specific mechanisms of antimalarial activity. Elife, 9. doi:10.7554/eLife.60246
- Strobel, H. A., LaBelle, S. A., Krishnan, L., Dale, J., Rauff, A., Poulson, A. M., Bader, N., Beare, J. E., Aliaj, K., Weiss, J. A., & Hoying, J. B. (2020). Stromal Cells Promote Neovascular Invasion Across Tissue Interfaces. Front Physiol, 11, 1026. doi:10.3389/fphys.2020.01026
2021
- de Hart, N. M. M.P.,Mahmassani, Z. S.,Reidy, P. T.,Kelley, J. J.,McKenzie, A. I.,Petrocelli, J. J.,Bridge, M. J.,Baird, L. M.,Bastian, E. D.,Ward, L. S.,Howard, M. T., & Drummond, M. J. (2021). Acute Effects of Cheddar Cheese Consumption on Circulating Amino Acids and Human Skeletal Muscle. Nutrients, 13(2). doi:10.3390/nu13020614
- Derksen, A., Shih, H.-Y., Forget, D., Darbelli, L., Tran, L. T., Poitras, C., . . . Bernard, G. (2021). Variants in LSM7 impair LSM complexes assembly, neurodevelopment in zebrafish and may be associated with an ultra-rare neurological disease. Human Genetics and Genomics Advances, 2(3), 100034. doi:https://doi.org/10.1016/j.xhgg.2021.100034
- Ferrara, P. J., Rong, X., Maschek, J. A., Verkerke, A. R., Siripoksup, P., Song, H., Green, T. D., Krishnan, K. C., Johnson, J. M., Turk, J., Houmard, J. A., Lusis, A. J., Drummond, M. J., McClung, J. M., Cox, J. E., Shaikh, S. R., Tontonoz, P., Holland, W. L., & Funai, K. (2021). Lysophospholipid acylation modulates plasma membrane lipid organization and insulin sensitivity in skeletal muscle. J Clin Invest, 131(8). doi:10.1172/JCI135963
- Ferrari, L., Pei, J., Zickella, M., Rey, C., Zickella, J., Ramirez, A., & Taylor, N. (2021). D2 Receptors in the Periaqueductal Gray/Dorsal Raphe Modulate Peripheral Inflammatory Hyperalgesia via the Rostral Ventral Medulla. Neuroscience, 463, 159-173. doi:10.1016/j.neuroscience.2021.03.035
- Grow, E. J., Weaver, B. D., Smith, C. M., Guo, J., Stein, P., Shadle, S. C., Hendrickson, P. G., Johnson, N. E., Butterfield, R. J., Menafra, R., Kloet, S. L., van der Maarel, S. M., Williams, C. J., & Cairns, B. R. (2021). p53 convergently activates Dux/DUX4 in embryonic stem cells and in facioscapulohumeral muscular dystrophy cell models. Nat Genet. doi:10.1038/s41588-021-00893-0
- Lewis, M. (2021). Signaling Pathway Regulation of Transcription Factor E2F1 Alters Cell Cycle Progression in Drosophila Midgut. In B. Edgar & J. I. Øvrebø (Eds.), (Vol. 21). Undergraduate Research Journal: University of Utah.
- Lyu, Y., Verma, V. K., Lee, Y., Taleb, I., Badolia, R., Shankar, T. S., Kyriakopoulos, C. P., Selzman, C. H., Caine, W., Alharethi, R., Navankasattusas, S., Seidel, T., Drakos, S. G., & Sachse, F. B. (2021). Remodeling of t-system and proteins underlying excitation-contraction coupling in aging versus failing human heart. NPJ Aging Mech Dis, 7(1), 16. doi:10.1038/s41514-021-00066-7
- Mahmassani, Z. S., McKenzie, A. I., Petrocelli, J. J., de Hart, N. M., Reidy, P. T., Fix, D. K., Ferrara, P. J., Funai, K., & Drummond, M. J. (2021). Short-term metformin ingestion by healthy older adults improves myoblast function. Am J Physiol Cell Physiol, 320(4), C566-C576. doi:10.1152/ajpcell.00469.2020
- Mathew, B., Aoyagi, K. L., & Fisher, M. A. (2021). Yersinia pestis lipopolysaccharide remodeling confers resistance to a Xenopsylla cheopis cecropin. bioRxiv, 2021.2003.2012.435208. doi:10.1101/2021.03.12.435208
- Moriwaki, M., & Welt, C. K. (2021). PRL Mutation Causing Alactogenesis: Insights Into Prolactin Structure and Function Relationships. J Clin Endocrinol Metab, 106(8), e3021-e3026. doi:10.1210/clinem/dgab201
- Russell, K. L., Downie, J. M., Gibson, S. B., Tsetsou, S., Keefe, M. D., Duran, J. A., Figueroa, K. P., Bromberg, M. B., Murtaugh, L. C., Bonkowsky, J. L., Pulst, S. M., & Jorde, L. B. (2021). Pathogenic Effect of TP73 Gene Variants in People With Amyotrophic Lateral Sclerosis. Neurology. doi:10.1212/WNL.0000000000012285
- Tamamouna, V, Rahman, MM, Petersson, M, Charalambous, I, Kux, K, Mainor, H, Bolender, V, Isbilir, B, Edgar, BA, & Pitsouli, C. (2021). Remodeling of oxygen-transporting tracheoles drives intestinal regeneration and tumorigenesis in Drosophila. Nature Cell Biology, 23(5), 497-+. doi:10.1038/s41556-021-00674-1
- Wike, C. L., Guo, Y., Tan, M., Nakamura, R., Shaw, D. K., Díaz, N., Whittaker-Tademy, A. F., Durand, N. C., Aiden, E. L., Vaquerizas, J. M., Grunwald, D., Takeda, H., & Cairns, B. R. (2021). Chromatin architecture transitions from zebrafish sperm through early embryogenesis. Genome Res, 31(6), 981-994. doi:10.1101/gr.269860.120
- Zhang, P., Katzaroff, A. J., Buttitta, L. A., Ma, Y., Jiang, H., Nickerson, D. W., Øvrebø, J. I., & Edgar, B. A. (2021). The Krüppel-like factor Cabut has cell cycle regulatory properties similar to E2F1. Proc Natl Acad Sci U S A, 118(7). doi:10.1073/pnas.2015675118
2019
- Deshpande, I., Liang, J., Hedeen, D., Roberts, K. J., Zhang, Y., Ha, B., Latorraca, N. R., Faust, B., Dror, R. O., Beachy, P. A., Myers, B. R., & Manglik, A. (2019). Smoothened stimulation by membrane sterols drives Hedgehog pathway activity. Nature, 571(7764), 284-+. doi:10.1038/s41586-019-1355-4
- Feng, H. D., Magda, J. J., & Gale, B. K. (2019). Viscoelastic second normal stress difference dominated multiple-stream particle focusing in microfluidic channels. Appl Phys Lett, 115(26), 263702(1-5) doi:Artn 26370210.1063/1.5129281
- Hanak, T. J., Libbey, J. E., Doty, D. J., Sim, J. T., DePaula-Silva, A. B., & Fujinami, R. S. (2019). Positive modulation of mGluR5 attenuates seizures and reduces TNF-alpha(+) macrophages and microglia in the brain in a murine model of virus-induced temporal lobe epilepsy. Exp Neurol, 311, 194-204. doi:10.1016/j.expneurol.2018.10.006
- Kim, H. S., Neugebauer, J., McKnite, A., Tilak, A., & Christian, J. L. (2019). BMP7 functions predominantly as a heterodimer with BMP2 or BMP4 during mammalian embryoaenesis. Elife, 8. doi:ARTN e48872 10.7554/eLife.48872
- Reidy, P. T., Yonemura, N. M., Madsen, J. H., McKenzie, A. I., Mahmassani, Z. S., Rondina, M. T., Lin, Y. K., Kaput, K., & Drummond, M. J. (2019). An accumulation of muscle macrophages is accompanied by altered insulin sensitivity after reduced activity and recovery. Acta Physiol, 226(2). doi:UNSP e13251 10.1111/apha.13251
2020
- Dong, Z. M., Lin, E., Wechsler, M. E., Weller, P. F., Klion, A. D., Bochner, B. S., Delker, D. A., Hazel, M. W., Fairfax, K., Khoury, P., Akuthota, P., Merkel, P. A., Dyer, A. M., Langford, C., Specks, U., Gleich, G. J., Chinchilli, V. M., Raby, B., Yandell, M., & Clayton, F. (2020). Pulmonary Eosinophilic Granulomatosis with Polyangiitis Has IgG4 Plasma Cells and Immunoregulatory Features. Am J Pathol, 190(7), 1438-1448. doi:10.1016/j.ajpath.2020.03.005
- Feng, H. D., Hockin, M., Zhang, S. H., Capecchi, M., Gale, B., & Sant, H. (2020). Enhanced chromosome extraction from cells using a pinched flow microfluidic device. Biomed Microdevices, 22(2). doi:ARTN 25 10.1007/s10544-020-0477-7
- Gandelman, M., Dansithong, W., Figueroa, K. P., Paul, S., Scoles, D. R., Pulst, S. M. (2020). Staufen 1 amplifies proapoptotic activation of the unfolded protein response. Cell Death Differ. doi:10.1038/s41418-020-0553-9
- Guo,P., Nalder,S., Okada,M., Sigala,A.P. (2020). Doxycycline has Distinct Apicoplast-Specific Mechanisms of Antimalarial Activity. BioRxiv 2020.06.11.146407; doi: https://doi.org/10.1101/2020.06.11.146407
- Hoffman, L.M., Smith, M.A., Jensen, C.C., Yoshigi, M. , Blankman, E., Ullman, K., Beckerle, M.C., (2020). Mechanical Stress Triggers Nuclear Remodeling and the Formation of Transmembrane Actin Nuclear Lines With Associated Nuclear Pore Complexes. Mol Biol Cell., DOI: 10.1091/mbc.E19-01-0027
- Lim, K., Sima, M., Stewart, R. J., & Minteer, S. D. (2020). Direct bioelectrocatalysis by redox enzymes immobilized in electrostatically condensed oppositely charged polyelectrolyte electrode coatings. Analyst, 145(4), 1250-1257. doi:10.1039/c9an02168j
- Mahmassani, Z. S., Reidy, P. T., McKenzie, A. I., Petrocelli, J. J., Matthews, O., de Hart, N. M., Ferrara, P. J., O'Connell, R. M., Funai, K., & Drummond, M. J. (2020). Absence of MyD88 from Skeletal Muscle Protects Female Mice from Inactivity-Induced Adiposity and Insulin Resistance. Obesity, 28(4), 772-782. doi:10.1002/oby.22759
- McKenzie, A. I., Reidy, P. T., Nelson, D. S., Mulvey, J. L., Yonemura, N. M., Petrocelli, J. J., Mahmassani, Z. S., Tippetts, T. S., Summers, S. A., Funai, K., & Drummond, M. J. (2020). Pharmacological inhibition of TLR4 ameliorates muscle and liver ceramide content after disuse in previously physically active mice. Am J Physiol-Reg I, 318(3), R503-R511. doi:10.1152/ajpregu.00330.2019
- Than, H., Pomicter, A. D., Yan, D., Beaver, L. P., Eiring, A. M., Heaton, W. L., Senina, A., Clair, P. M., Shacham, S., Mason, C. C., Hare, T. O., & Deininger, M. W. (2020). Coordinated inhibition of nuclear export and Bcr-Abl1 selectively targets chronic myeloid leukemia stem cells. Leukemia, 34(6), 1679-1683. doi:10.1038/s41375-020-0708-1


.svg%2F2560px-YouTube_full-color_icon_(2017).svg.png&f=1&nofb=1&ipt=73e306e7b63e24993223ed90fa2c25326736677bb7eb6f01e1c9885abf904c81&ipo=images)