
Mass Spectrometry and Proteomics Core
The mission of the Proteomics Core at the University of Utah is to provide expert proteomics services and consultation for the University of Utah research communities, as well as to public and private institutions within the state of Utah. The goals of the facility are to provide access to high performance mass spectrometry resources and the latest proteomics technology for researchers in addition to supporting an efficient research and educational environment for sharing ideas, experiences, and knowledge of proteomics. We strive to provide the highest quality mass spectrometry analyses for protein investigations.
The Proteomics Core enables investigators to utilize an array of state-of-the-art mass spectrometry-based techniques for proteomics experiments, ranging from protein identification, post-translational modification characterizations and protein interactions to large-scale quantitative proteomic analyses using data-independent acquisition and tandem mass tag labeling. The staff is available to users for consultation on experimental design and proteomics methods, assisting with data interpretation, manuscript preparation and grant submission.
The primary focus of the Proteomics Core is to support excellent proteomics research in academic, clinical and industry settings. A new project consultation meeting with a proteomics facility core staff member is required for each new project. Please contact the proteomics core facility staff to arrange a consultation meeting prior to sample submission. This core provides and operates as a fee-for-service resource with routine services listed below. Common reagents necessary to perform each proteomics experiment (e.g., reducing and alkylating agents, trypsin, Ziptips, IMAC enrichment kits, tandem mass tags and LC/MS grade solvents) are provided by the Proteomics Core and the costs are already built into the estimated service fee. All project costs will fluctuate based on sample size and experimental design. We will provide a cost estimate following a project consultation with a staff member prior to sample submission.
A new project consultation meeting with a proteomics facility core staff member is required for each new project. Please contact the proteomics core facility staff to arrange a consultation meeting prior to sample submission.
Major
Instruments
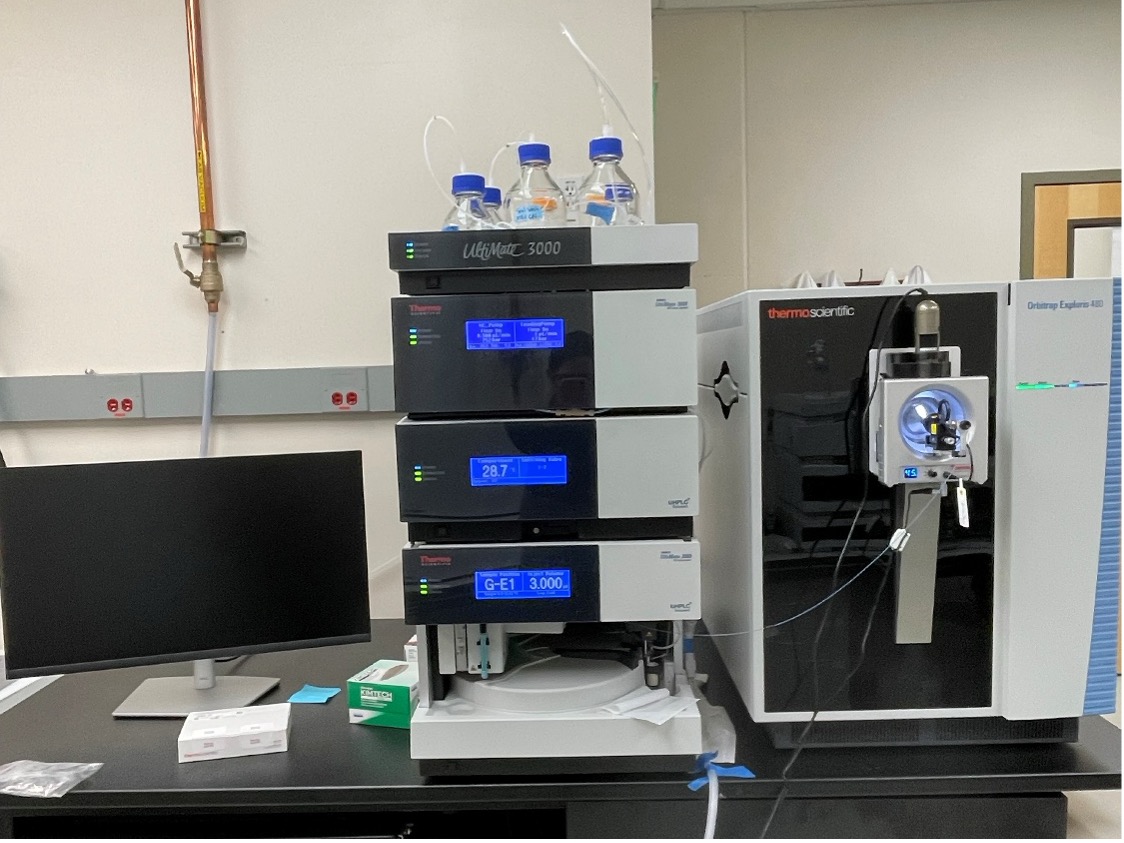
Orbitrap Exploris™ 480
The Orbitrap Exploris™ 480 Mass Spectrometer (ThermoFisher Scientific) is coupled to a Dionex Ultimate3000 RSLCnano System and nano-electrospray ionization for ultimate sensitivity and chromatographic performance. Especially well-suited for proteomics applications, the Exploris 480 instrument provides high sensitivity (sub-femtomole), high resolution up to 480,000, and high mass accuracy (< 2 ppm), affording greater sequence information and high-confidence protein identifications. The Exploris 480 is well equipped for protein identification, mapping protein post-translational modifications and protein quantification via tandem-mass tag labeling.

Maxis II ETD
The Maxis II ETD (Bruker) is paired with an Ekspert nanoLC 400 liquid chromatography system and electrospray ionization source. The Maxis is optimal for providing high mass accuracy for intact protein analysis.

timsTOF Pro 2
The timsTOF Pro2 mass spectrometer (Bruker) is coupled to a nanoElute2 nano liquid chromatography system (Bruker) that is advantageous for high throughput label-free quantification of the whole proteome. Trapped ion mobility spectrometry (TIMS) is a gas phase separation technique that resolves sample complexity through an added dimension of separation improving the number of identified peptides in a whole cell or tissue lysate. Time of flight (TOF) is a mass analyzer that uses an electric field to accelerate generated ions through the same electrical potential, and then measures the time each ion takes to reach the detector with high mass accuracy (< 3 ppm) eliciting high confidence peptide identifications.

Data Analysis Software
- Proteome Discoverer 3.0 coupled with SEQUEST and MASCOT
- Thermo Scientific Xcalibur
- Skyline 21.2
- Bruker PaSER version (2023b)
- DIA-NN v1.8.1
- MSfragger v20.0
Available Services
Common reagents necessary to perform each proteomics experiment (e.g., reducing and alkylating agents, trypsin, Ziptips, IMAC enrichment kits, tandem mass tags and LC/MS grade solvents) are provided by the Proteomics Core and the costs are already built into the estimated service fee. All project costs will fluctuate based on sample size and experimental design. We will provide a cost estimate following a project consultation with a staff member prior to sample submission.
Please consult the FAQs page or contact a staff member if you have any questions about our services.
General Services
- Intact Mass Analysis of proteins or small biomolecules
- Electrospray Ionization (ESI) Direct Sample Injection
Protein Identification
- Targeted and untargeted protein identification
- Identification of protein-protein interacting partners
- Peptide enrichment and identification of post-translational modifications (PTMs)
- High pH Fractionation: Increases protein identification from LC-MS/MS analysis through orthogonal peptide fractionation of complex peptide samples
Quantitative Proteomics
- Data-independent acquisition (DIA) label-free
- Tandem Mass Tags (TMT) labeling
- Stable Isotope Labeling by Amino acids in Cell culture (SILAC)
Proteomics LC-MS/MS Mass Spectrometry Project Rates
Internal Academic (University of Utah)
| Sample Preparation | Instrument run time | Data Analysis |
| $110/ hour + cost of supplies | $90.00/ run | $110.00/ hour |
External Academic (outside of University of Utah)
| Sample Preparation | Instrument run time | Data Analysis |
| $165/ hour + cost of supplies | $135.00/ run | $165.00/ hour |
Commercial / Industry Customers
| Sample Preparation | Instrument run time | Data Analysis |
| $220/ hour + cost of supplies | $180.00/ run | $220.00/ hour |
* The cost of supplies is based on sample size and experimental design
* The Proteomics Core Facility will provide a cost estimate that reflects individual experimental design and sample size prior to sample submission
Intact Mass Analysis Mass Spectrometry Project Rates
Internal Academic (University of Utah)
| Direct Inject | LC-MS | Data Analysis |
| $80.00 / sample | $184.00 / sample | $110.00/ hour |
External Academic (outside of University of Utah)
| Direct Inject | LC-MS | Data Analysis |
| $120.00 / sample | $276.00 / sample | $165.00/ hour |
Commercial / Industry Customers
| Direct Inject | LC-MS | Data Analysis |
| $190.00 / sample | $368.00 / sample | $220.00/ hour |
Project Rates
The Proteomics Core Facility will provide a cost estimate that reflects individual experimental design and sample size prior to sample submission.
University of Utah Internal Customers
| Service | Lab Supplies (per sample) | Scientist Time (per hour) | Instrument Time (per sample) | Data Analysis (per hour) |
| Protein Identification | $30.00 | $100.00 | $80.00 | $100.00 |
| Protein Quantification (DIA) | $30.00 | $100.00 | $80.00 | $100.00 |
| Protein Quantification (TMT10plex kit) | $650.00 | $100.00 | $80.00 | $100.00 |
| High pH Fractionation (8 fractions/sample) | $60.00 | $100.00 | $640.00 | $100.00 |
| ESI Direct Injection | - | $100.00 | $22.00 | $100.00 |
| Intact Mass Analysis | - | $100.00 | $55.00 | $100.00 |
| PTM Enrichment (e.g., Phosphoproteomics) | $472.50 | $100.00 | $80.00 | $100.00 |
Academic External Customers
| Service | Lab Supplies (per sample) | Scientist Time (per hour) | Instrument Time (per sample) | Data Analysis (per hour) |
| Protein Identification | $45.00 | $150.00 | $120.00 | $150.00 |
| Protein Quantification (DIA) | $45.00 | $150.00 | $120.00 | $150.00 |
| Protein Quantification (TMT10plex kit) | $975.00 | $150.00 | $120.00 | $150.00 |
| High pH Fractionation (8 fractions/sample) | $90.00 | $150.00 | $960.00 | $150.00 |
| ESI Direct Injection | - | $150.00 | $33.00 | $150.00 |
| Intact Mass Analysis | - | $150.00 | $83.00 | $150.00 |
| PTM Enrichment (e.g., Phosphoproteomics) | $708.00 | $150.00 | $120.00 | $150.00 |
Commercial / Industry Customers
| Service | Lab Supplies (per sample) | Scientist Time (per hour) | Instrument Time (per sample) | Data Analysis (per hour) |
| Protein Identification | $60.00 | $200.00 | $160.00 | $200.00 |
| Protein Quantification (DIA) | $60.00 | $200.00 | $160.00 | $200.00 |
| Protein Quantification (TMT10plex kit) | $1300.00 | $200.00 | $160.00 | $200.00 |
| High pH Fractionation (8 fractions/sample) | $120.00 | $200.00 | $1280.00 | $200.00 |
| ESI Direct Injection | - | $200.00 | $44.00 | $200.00 |
| Intact Mass Analysis | - | $200.00 | $165.00 | $200.00 |
| PTM Enrichment (e.g., Phosphoproteomics) | $945.00 | $200.00 | $160.00 | $200.00 |
Requesting Services
Existing users may login directly to the Resource Scheduling System to schedule or order services. This system is cores-wide and uses University of Utah uNID authentication.
Contact Us
Hours of Operation
9:00 am to 5:00 pm
Monday - Friday
Shipping Address
University of Utah
Mass Spectrometry & Proteomics Core
15 N Medical Drive East, RM A306
Salt Lake City, UT 84112-5650 USA
Major Acknowledgements
- Cox HD, Smeal SJ, Hughes CM, Cox JE, Eichner D. Detection and in vitro metabolism of AOD9604. Drug Test Anal. 2015;7(1):31-8. Epub 2014/09/12. doi: 10.1002/dta.1715. PubMed PMID: 25208511.
- Shen PS, Park J, Qin Y, Li X, Parsawar K, Larson MH, Cox J, Cheng Y, Lambowitz AM, Weissman JS, Brandman O, Frost A. Protein synthesis. Rqc2p and 60S ribosomal subunits mediate mRNA-independent elongation of nascent chains. Science. 2015;347(6217):75-8. Epub 2015/01/03. doi: 10.1126/science.1259724. PubMed PMID: 25554787; PMCID: PMC4451101.
Recent Acknowledgements
- Espino-Sanchez TJ, Wienkers H, Marvin RG, Nalder SA, Garcia-Guerrero AE, VanNatta PE, Jami-Alahmadi Y, Mixon Blackwell A, Whitby FG, Wohlschlegel JA, Kieber-Emmons MT, Hill CP, Sigala PA. Direct tests of cytochrome c and c(1) functions in the electron transport chain of malaria parasites. Proc Natl Acad Sci U S A. 2023;120(19):e2301047120. Epub 2023/05/01. doi: 10.1073/pnas.2301047120. PubMed PMID: 37126705; PMCID: PMC10175771.
- Luke EN, Ratnatilaka Na Bhuket P, Yu SM, Weiss JA. Targeting damaged collagen for intra-articular delivery of therapeutics using collagen hybridizing peptides. J Orthop Res. 2023. Epub 2023/04/23. doi: 10.1002/jor.25577. PubMed PMID: 37087677.
- Subrahmanyam N, Yathavan B, Kessler J, Yu SM, Ghandehari H. HPMA copolymer-collagen hybridizing peptide conjugates targeted to breast tumor extracellular matrix. J Control Release. 2023;353:278-88. Epub 2022/10/17. doi: 10.1016/j.jconrel.2022.10.017. PubMed PMID: 36244509.
- Wenzel DM, Mackay DR, Skalicky JJ, Paine EL, Miller MS, Ullman KS, Sundquist WI. Comprehensive analysis of the human ESCRT-III-MIT domain interactome reveals new cofactors for cytokinetic abscission. Elife. 2022;11. Epub 2022/09/16. doi: 10.7554/eLife.77779. PubMed PMID: 36107470; PMCID: PMC9477494.
Citing Our Facility
Acknowledgments
We would like to thank you for acknowledging the our facility. This recognition allows us to highlight the impact of your work and demonstrates the important contributions of our facility makes to research across the University of Utah. The recognition our core receives from your acknowledgments also aids in receiving grants and further funding for equipment and services we can provide to our users.
Self-Run Services / Instrumentation Usage:
In published papers that used instruments at our facility and notably involved staff members please use the following format:
We acknowledge (facility name) at the University of Utah for use of equipment (insert instrument/service details here), and thank (insert any notable staff member – if desired) for their assistance.
Assisted Services:
In published papers where a staff member assisted you in addition to the requested services please use the following format:
We acknowledge (facility name) at the University of Utah for use of equipment (insert instrument/service details here), and thank (insert staff member-required) for their assistance in (service provided).
Collaboration:
For publications resulting from collaborations that assisted with the methodologies, planning process and execution of your experiment in addition to equipment usage we require Co-author attribution on your publication for our facility and any staff members who provided substantial contributions to the originating project.







