
DNA/Peptide Synthesis Core
The DNA/Peptide Synthesis Core provides researchers with solid-phase chemical synthesis of oligonucleotides and peptides. Services offered include a variety of synthesis scales and purity options to meet a wide range of experimental needs. In addition to synthesis with standard bases and amino acids, the facility also offers hundreds of specialty modifications that can be incorporated. Some common examples include phosphorylation, biotinylation, flours and fluor/quencher pairs, as well as several types modified-monomers. Many investigators also utilize functional group modifications such as amino, thiol, or alkyne/azide groups for use in conjugation or immobilization reactions.
Our on-campus location allows for rapid turn-around times and convenient delivery of products. Feel free to contact us with any questions or have a look at our FAQs, where you’ll find answers to many of the more common ones.
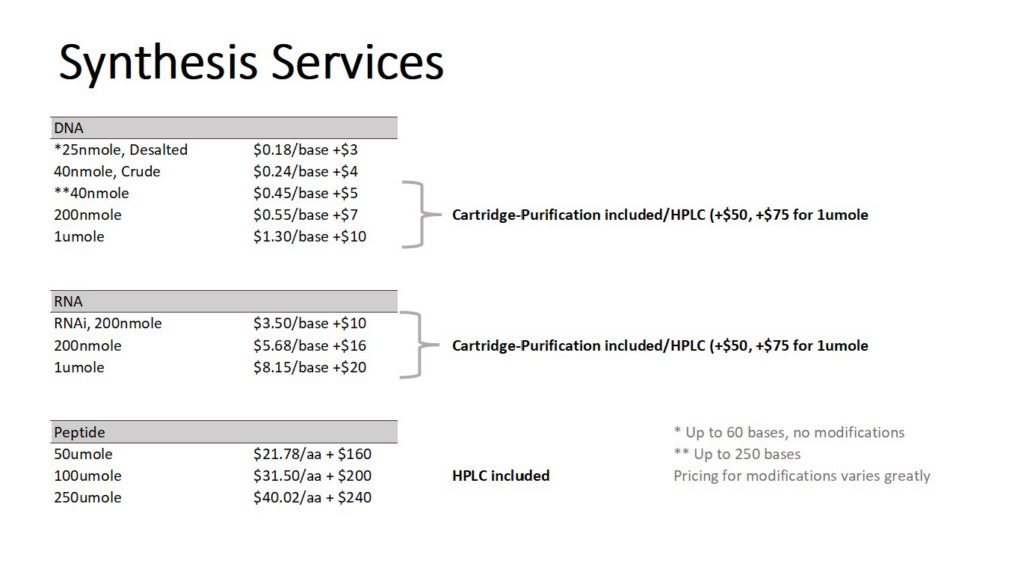
Service Rates
Requesting Services
Existing users may login directly to the Resource Scheduling System to schedule or order services. This system is cores-wide and uses University of Utah uNID authentication.
Screening Attestation
US Framework for Nucleic Acid Synthesis Screening
The DNA/Peptide Synthesis HSC Core Facility understands that United States federally funded life sciences entities and funding agencies are relying on this attestation prior to purchasing synthetic oligonucleotides. The DNA/Peptide Synthesis HSC Core Facility self-attests to complying with the US Framework for Nucleic Acid Synthesis Screening. Should this information change, the DNA/Peptide Synthesis HSC Core Facility agrees that it will update this statement within 72 hours. For more information, please contact Mike Hanson.
RNA
What kinds of purifications are available?
Most RNA oligos are either cartridge-purified or HPLC-purified. Specialty modifications, however, can preclude the ability for cartridge-purification. In such cases, we provide desalted product.
How should I store my RNA? Is it stable?
The best way to store RNA is to dry the oligo from a solution of water either by evaporation or precipitation. We provide the RNA in this dry form, and it should be stable for a year or more if kept at 4°C or lower. To store RNA in solution, the pH should be kept near neutral and nuclease-free. It’s best to keep it frozen in aliquots to minimize the number of freeze-thaw cycles. Under these conditions, RNA should be stable for at least 6 months in solution.
If I send you my target gene sequence, can you design my siRNAs for me?
Unfortunately, we do not design either RNA or DNA primers for users at this time. Please see our guidelines below if you are a first-time user ordering RNAi.
Guidelines for ordering RNAi
- All of our RNA synthesis services can be used to make siRNA. The “RNAi” service is distinct in that RNA synthesized in this manner must contain at least one 3′-deoxy overhang. The most common approach is to design sense and anti-sense 21mers, containing 19 RNA bases and 3′-dTdT overhangs. The 3′-deoxy overhangs allow for more efficient and less-costly synthesis (for 200nmole syntheses), and may enhance the efficacy of silencing.
- To maximize stability, we provide the sense and antisense RNAi lyophilized and in separate tubes. The user anneals the two complementary strands to form a duplex molecule.
- 200nmol “RNAi” syntheses generally give 20 -70 nmoles of cartridge-purified material.
Peptide
What about peptide quantitation options?
Weighing a peptide after lyophilization may result in errors of up to 50% because of varying amounts of water left in the powder. The amount of water is difficult to control, even when the drying has been done thoroughly. Better ways to measure peptides are:
- Amino acid analysis is the “gold standard” for measuring peptides. When you know the amino acid content, the test results in measurements of each amino acid present. AAA can be used for any peptide, regardless of amino acid content. Unfortunately, we do not offer this service.
- The Ellman reaction is a very reliable measurement of reduced –SH groups on cysteine. If a peptide has a cysteine, we will provide amounts in the fraction table based on Ellman measurement. The reaction is easily done in the user’s lab as well. Ask us for a procedure.
- The UV absorbance of peptides containing Tyrosine, and/or Tryptophan can be measured, and a theoretical extinction coefficient calculated. This is also easily done in the user’s lab.
Is this a difficult peptide?
Peptides vary considerably in their difficulty to synthesize and purify. There are too many variables to describe here. Core facility chemists will be happy to advise users of foreseeable difficulties and suggest options. Please call to discuss or request quotations for complex syntheses.
How long will it take?
We try to deliver peptides within three weeks after they are ordered. Since peptide synthesis is unpredictable we may need to repeat a synthesis to get the purity you need. If there is a backlog at the time of ordering, we will inform you of likely delays.
Major Acknowledgements
- Hanson WM, Chen Z, Jackson LK, Attaf M, Sewell AK, Heemstra JM, Phillips JD. Reversible Oligonucleotide Chain Blocking Enables Bead Capture and Amplification of T-Cell Receptor α and β Chain mRNAs. J Am Chem Soc. 2016 Sep 7;138(35):11073-6. doi: 10.1021/jacs.6b04465. Epub 2016 Aug 1. PMID: 27478996; PMCID: PMC5249220.
Recent Acknowledgements
- Aderounmu AM, Aruscavage PJ, Kolaczkowski B, Bass BL. Ancestral protein reconstruction reveals evolutionary events governing variation in Dicer helicase function. Elife. 2023 Apr 17;12:e85120. doi: 10.7554/eLife.85120.
- Batistatou N, Kritzer JA. Investigation of Sequence-Penetration Relationships of Antisense Oligonucleotides. Chembiochem. 2023 May 2;24(9):e202300009. doi: 10.1002/cbic.202300009. Epub 2023 Apr 4.
- Bauer KM, Nelson MC, Tang WW, Chiaro TR, Brown DG, Ghazaryan A, Lee SH, Weis AM, Hill JH, Klag KA, Tran VB, Thompson JW, Ramstead AG, Monts JK, Marvin JE, Alexander M, Voth WP, Stephens WZ, Ward DM, Petrey AC, Round JL, O'Connell RM. CD11c+ myeloid cell exosomes reduce intestinal inflammation during colitis. JCI Insight. 2022 Oct 10;7(19):e159469. doi: 10.1172/jci.insight.159469.
- Campbell RA, Manne BK, Banerjee M, Middleton EA, Ajanel A, Schwertz H, Denorme F, Stubben C, Montenont E, Saperstein S, Page L, Tolley ND, Lim DL, Brown SM, Grissom CK, Sborov DW, Krishnan A, Rondina MT. IFITM3 regulates fibrinogen endocytosis and platelet reactivity in nonviral sepsis. J Clin Invest. 2022 Dec 1;132(23):e153014. doi: 10.1172/JCI153014.
- Fleming AM, Burrows CJ. Nanopore sequencing for N1-methylpseudouridine in RNA reveals sequence-dependent discrimination of the modified nucleotide triphosphate during transcription. Nucleic Acids Res. 2023 Feb 28;51(4):1914-1926. doi: 10.1093/nar/gkad044.
- Fleming AM, Tran R, Omaga CA, Howpay Manage SA, Burrows CJ, Conboy JC. Second Harmonic Generation Interrogation of the Endonuclease APE1 Binding Interaction with G-Quadruplex DNA. Anal Chem. 2022 Nov 1;94(43):15027-15032. doi: 10.1021/acs.analchem.2c02951. Epub 2022 Oct 21.
- Ghazaryan A, Wallace JA, Tang WW, Barba C, Lee SH, Bauer KM, Nelson MC, Kim CN, Stubben C, Voth WP, Rao DS, O'Connell RM. miRNA-1 promotes acute myeloid leukemia cell pathogenesis through metabolic regulation. Front Genet. 2023 May 9;14:1192799. doi: 10.3389/fgene.2023.1192799. PMID: 37229187; PMCID: PMC10203238.
- Howpay Manage SA, Zhu J, Fleming AM, Burrows CJ. Promoters vs. telomeres: AP-endonuclease 1 interactions with abasic sites in G-quadruplex folds depend on topology. RSC Chem Biol. 2023 Jan 18;4(4):261-270. doi: 10.1039/d2cb00233g.
- Howpay Manage SA, Fleming AM, Chen HN, Burrows CJ. Cysteine Oxidation to Sulfenic Acid in APE1 Aids G-Quadruplex Binding While Compromising DNA Repair. ACS Chem Biol. 2022 Sep 16;17(9):2583-2594. doi: 10.1021/acschembio.2c00511. Epub 2022 Aug 29.
- Manuel BA, Das S, Sanford A, Jenkins MC, Finn MG, Heemstra JM. Modular Catalysis: Aptamer Enhancement of Enzyme Kinetics in a Nanoparticle Reactor. Biomacromolecules. 2023 Apr 10;24(4):1934-1941. doi: 10.1021/acs.biomac.3c00144. Epub 2023 Mar 29.
- Montoya AL, Glavatskikh M, Halverson BJ, Yuen LH, Schüler H, Kireev D, Franzini RM. Combining pharmacophore models derived from DNA-encoded chemical libraries with structure-based exploration to predict Tankyrase 1 inhibitors. Eur J Med Chem. 2023 Jan 15;246:114980. doi: 10.1016/j.ejmech.2022.114980. Epub 2022 Dec 2.
- Myres GJ, Harris JM. Nanomolar Binding of an Antibiotic Peptide to DNA Measured with Raman Spectroscopy. Langmuir. 2023 Mar 21;39(11):4150-4160. doi: 10.1021/acs.langmuir.3c00099. Epub 2023 Mar 8.
- Myres GJ, Harris JM. Stable Immobilization of DNA to Silica Surfaces by Sequential Michael Addition Reactions Developed with Insights from Confocal Raman Microscopy. Anal Chem. 2023 Feb 14;95(6):3499-3506. doi: 10.1021/acs.analchem.2c05594. Epub 2023 Jan 31.
- Smith MA, Blankman E, Jensen CC, Hoffman LM, Ullman KS, Beckerle MC. Nuclear pore complexes concentrate on Actin/LINC/Lamin nuclear lines in response to mechanical stress in a SUN1 dependent manner. Heliyon. 2022 Dec 7;8(12):e12147. doi: 10.1016/j.heliyon.2022.e12147.
- Wenzel, D. M., D. R. Mackay, J. J. Skalicky, E. L. Paine, M. S. Miller, K. S. Ullman and W. I. Sundquist (2022). Comprehensive analysis of the human ESCRT-III-MIT domain interactome reveals new cofactors for cytokinetic abscission. Elife 11.10.7554/eLife.77779
- Winter, J. M., H. L. Fresenius, C. N. Cunningham, P. Wei, H. R. Keys, J. Berg, A. Bott, T. Yadav, J. Ryan, D. Sirohi, S. R. Tripp, P. Barta, N. Agarwal, A. Letai, D. M. Sabatini, M. L. Wohlever and J. Rutter (2022). Collateral deletion of the mitochondrial AAA+ ATPase ATAD1 sensitizes cancer cells to proteasome dysfunction. Elife 11.10.7554/eLife.82860
Citing Our Facility
Acknowledgments
We would like to thank you for acknowledging the our facility. This recognition allows us to highlight the impact of your work and demonstrates the important contributions of our facility makes to research across the University of Utah. The recognition our core receives from your acknowledgments also aids in receiving grants and further funding for equipment and services we can provide to our users.
Self-Run Services / Instrumentation Usage:
In published papers that used instruments at our facility and notably involved staff members please use the following format:
We acknowledge (facility name) at the University of Utah for use of equipment (insert instrument/service details here), and thank (insert any notable staff member – if desired) for their assistance.
Assisted Services:
In published papers where a staff member assisted you in addition to the requested services please use the following format:
We acknowledge (facility name) at the University of Utah for use of equipment (insert instrument/service details here), and thank (insert staff member-required) for their assistance in (service provided).
Collaboration:
For publications resulting from collaborations that assisted with the methodologies, planning process and execution of your experiment in addition to equipment usage we require Co-author attribution on your publication for our facility and any staff members who provided substantial contributions to the originating project.




