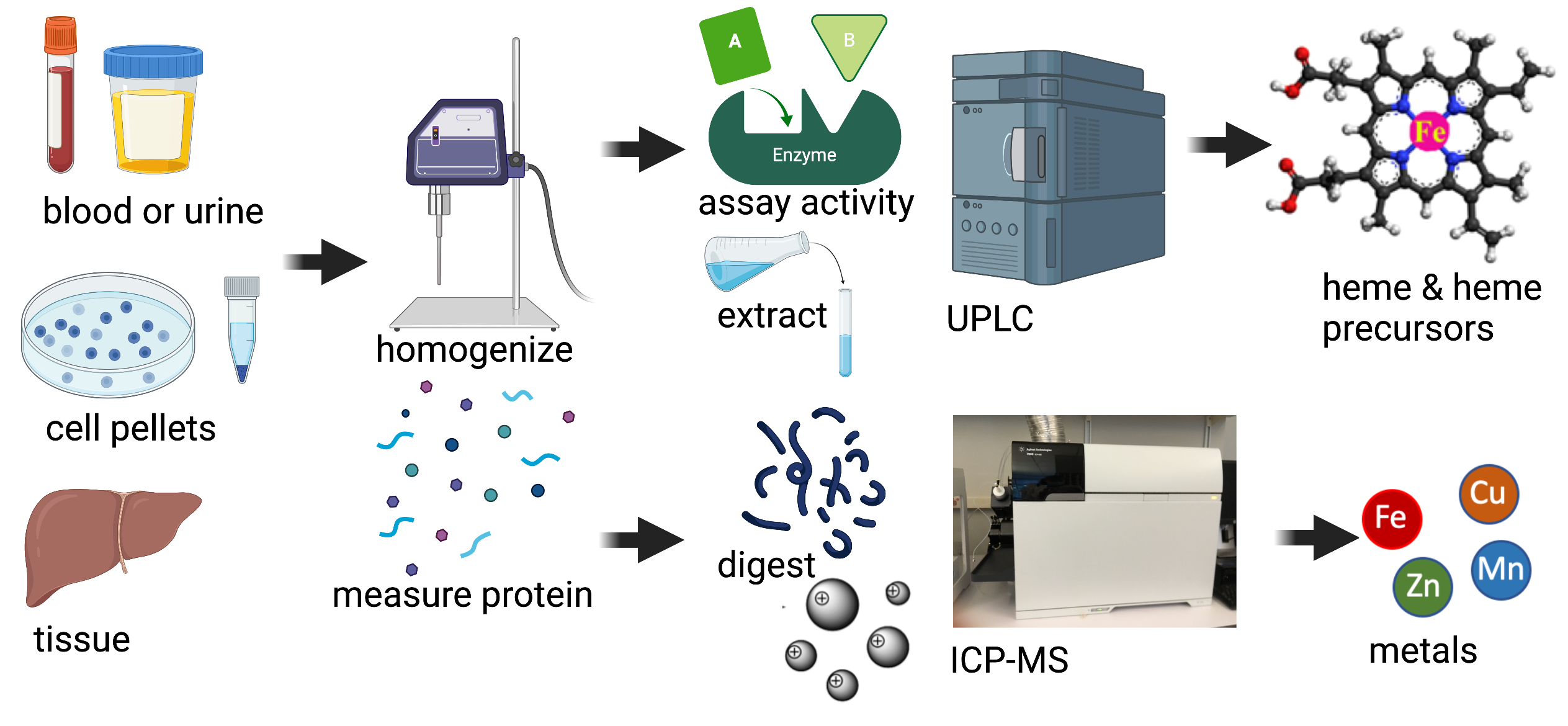
Iron and Heme Core Facility
The Iron and Heme Core Facility has been in operation since 2016 and is part of the NIDDK sponsored Center for Iron and Heme Disorders (NIH U54DK110858). It is also serves as one of the University of Utah shared-resource Health Science Cores facilities which are established service/recharge centers offering services to research facilities across the country. The Iron and Heme Core provides analyses of heme and precursor compounds and metals present in biological samples. The core specializes in the UPLC quantification of heme and other tetrapyrroles, and the measurement of activities of all eight enzymes in the heme biosynthetic pathway, and in the ICP-MS analysis of iron and other common metals in biological systems.
This Facility is equipped mainly with two Waters Acquity ultra performance liquid chromatography (UPLC) systems (Classic and H-Class Plus), each with its own solvent manager, sample manager, tandem high sensitivity photodiode array and fluorescence detectors, and reverse phase C18 column. An Agilent 7900 ICP-mass spectrometer with an SPS4 auto sampler is dedicated for metal analysis. Other supporting equipment allow for sample storage, handling, processing for analysis and data processing, and some of these are a dark room, fume hoods, a deoxygenation system, refrigeration and freezer space, a spectrophotometer, centrifuges, various tissue and cell disruptors, hot blocks, mixers and sample dryers.
With more than a quarter century of performing laboratory research on the heme biosynthetic pathway, the people with the Iron and Heme Core have contributed to high impact research that has lead to publications in PNAS, Elife, J. Biol. Chem., MicrobiologyOpen, Chemical Reviews, and in Blood Cells, Molecules, and Diseases.
Project Rates (as of July 1st, 2025)
| Service name | Sample Count | CIHD run setup fee | CIHD per sample charge | Non-CIHD run setup fee | Non-CIHD per sample charge |
| Heme, PPIX & ZnPPIX* | 1 - 24 | 395.00 | - | 1020.00 | - |
| Heme, PPIX & ZnPPIX, extra run required* | 1 - 24 | 295.00 | - | 610.00 | - |
| intermediate porphyrins | 1 - 24 | 350.00 | - | 980.00 | - |
| intermediate porphyrin isomers | 1 - 24 | 350.00 | - | 980.00 | - |
| Spectral analysis of hemes (pyridine hemochrome) | 1 - 24 | 60.00 | - | 1150.00 | - |
| ALA (aminolevulinic acid) assay | 1 - 12 | 340.00 | - | 960.00 | - |
| PBG (porphobilinogen) assay | 1- 12 | 340.00 | - | 960.00 | - |
| ALAS enzyme assay | 1 - 12 | 460.00 | - | 1700.00 | - |
| ALAD enzyme assay | 1- 12 | 340.00 | - | 1590.00 | - |
| PBGD enzymeassay | 1 - 12 | 340.00 | - | 1590.00 | - |
| U3S enzyme assay | 1- 12 | 340.00 | - | 1590.00 | - |
| UROD enzyme assay | 1 - 12 | 340.00 | - | 1590.00 | - |
| CPOX enzyme assay | 1- 12 | 420.00 | - | 1680.00 | - |
| PPOX enzyme assay | 1- 12 | 420.00 | - | 1680.00 | - |
| FECH enzyme assay | 1 - 12 | 375.00 | - | 1630.00 | - |
| Reverse FECH assay | 1- 12 | 420.00 | - | 1680.00 | - |
| ICP-MS metals | 1 - 36 | 490.00 | 2.50 | 925.00 | 2.50 |
| BCA assay (iron) per 12 | 1- 12 | 17.00 | - | 125.00 | - |
| Tissue homogenization (bead tubes)** | 1 | 6.00 | 8.70 |
* Only two components may be quantified at most in the same run. If all three were requested, that would be two runs for the same set of samples. If samples do not allow quantification of two components in the same run (amounts present are very disparate), another run will be performed to complete the requested service. The extra run would require 1/2 of consumables, no BCA, and 1/2 tech time. (For example, blood and spleen have very high heme and need two runs to quantify heme and PPIX/ZnPPIX.)
** The listed pricing for an ICP-MS run is for one to five elements; every element beyond that incurs an added 5% charge increase to the price of the analytical run.
*** For analyses on all tissues not needing enzyme activity measurements.
Contact us for similar services not listed above.
Note for all people sending samples for analyses:
With the act of sending samples to the Iron and Heme Core Facility for analysis, the sender also attests that the said samples and the accompanying packaging do not contain any hazardous components and are not potentially infections. If these samples were ever infectious, they should have been rendered noninfectious and verified to be so before shipping.
Requesting Services
Existing users may login directly to the Resource Scheduling System to schedule or order services. This system is cores-wide and uses University of Utah uNID authentication.
Preparation of Samples for Shipping
For most assays please send the samples as follows:
| Sample Type | Minumum Quantity | Form/Condition |
| Serum, Body Fluids | 1 mL | Quickly Frozen |
| Cells | > 100 uL packed cell pallet | Washed with PBS, pelleted, supernatant discarded and then frozen |
| Tissue | ~ 100 mg | Flash Frozen |
| Fresh Blood | 1 standard collection tube | With anticoagulant, in ice bath, and covered with foil |
| Feed and other complex/hard materials | 1 g | dried and ground to fine powder |
- Please send all frozen samples packed in dry ice for overnight shipping and weekday delivery to:
Iron and Heme Core Facility
Wintrobe Research Building Rm #621
26 North Medical Drive
Salt Lake City, UT 84132
Notify the Iron & Heme Core to ensure that someone is available to receive the samples. - Please contact us for the quantity needed if you want us to run multiple assays per sample.
- Unless otherwise requested, results of analyses heme and its precursor compounds shall be reported in nano- or picomoles per mg protein in the sample, and enzyme activity shall be reported as nano- or picomoles product per mg of total sample protein per hour.
- Please notify us if you wish to receive leftover samples after processing and assay has been completed.
- A FedEx number is required if the project is for an external user.
- All samples will be discarded 30 days after results have been sent to the investigator’s lab, unless we receive a return request.
Hours of Operation
8:00 am to 5:00 pm
Monday - Friday
Mailing Address
Iron and Heme Core Facility
Wintrobe Research Building, Rm #621
26 North Medical Drive
Salt Lake City, UT 84132
Major Acknowledgements
- Chung J, Wittig JG, Ghamari A, Maeda M, Dailey TA, Bergonia H, Kafina MD, Coughlin EE, Minogue CE, Hebert AS, Li L, Kaplan J, Lodish HF, Bauer DE, Orkin SH, Cantor AB, Maeda T, Phillips JD, Coon JJ, Pagliarini DJ, Dailey HA, Paw BH. Erythropoietin signaling regulates heme biosynthesis. Elife. 2017 May 29;6:e24767. doi: 10.7554/eLife.24767. PMID: 28553927; PMCID: PMC5478267.
- Kusminski CM, Ghaben AL, Morley TS, Samms RJ, Adams AC, An Y, Johnson JA, Joffin N, Onodera T, Crewe C, Holland WL, Gordillo R, Scherer PE. A Novel Model of Diabetic Complications: Adipocyte Mitochondrial Dysfunction Triggers Massive β-Cell Hyperplasia. Diabetes. 2020 Mar;69(3):313-330. doi: 10.2337/db19-0327. Epub 2019 Dec 27. PMID: 31882562; PMCID: PMC7034182.
- Pek RH, Yuan X, Rietzschel N, Zhang J, Jackson L, Nishibori E, Ribeiro A, Simmons W, Jagadeesh J, Sugimoto H, Alam MZ, Garrett L, Haldar M, Ralle M, Phillips JD, Bodine DM, Hamza I. Hemozoin produced by mammals confers heme tolerance. Elife. 2019 Oct 1;8:e49503. doi: 10.7554/eLife.49503. Erratum in: Elife. 2023 Oct 20;12: PMID: 31571584; PMCID:
- Rocha ER, Bergonia HA, Gerdes S, Jeffrey Smith C. Bacteroides fragilis requires the ferrous-iron transporter FeoAB and the CobN-like proteins BtuS1 and BtuS2 for assimilation of iron released from heme. Microbiologyopen. 2019 Apr;8(4):e00669. doi: 10.1002/mbo3.669. Epub 2018 Jun 21. PMID: 29931811; PMCID: PMC6460266.
- Yien YY, Ducamp S, van der Vorm LN, Kardon JR, Manceau H, Kannengiesser C, Bergonia HA, Kafina MD, Karim Z, Gouya L, Baker TA, Puy H, Phillips JD, Nicolas G, Paw BH. Mutation in human CLPX elevates levels of δ-aminolevulinate synthase and protoporphyrin IX to promote erythropoietic protoporphyria. Proc Natl Acad Sci U S A. 2017 Sep 19;114(38):E8045-E8052. doi: 10.1073/pnas.1700632114. Epub 2017 Sep 5. PMID: 28874591; PMCID: PMC5617249.
Recent Acknowledgements
- Jackson LK, Dailey TA, Anderle B, Warren MJ, Bergonia HA, Dailey HA, Phillips JD. Exploiting Differences in Heme Biosynthesis between Bacterial Species to Screen for Novel Antimicrobials. Biomolecules. 2023 Oct 6;13(10):1485. doi: 10.3390/biom13101485. PMID: 37892169; PMCID: PMC10604556.
- Miljkovic M, Seguin A, Jia X, Cox JE, Catrow JL, Bergonia H, Phillips JD, Stephens WZ, Ward DM. Loss of the mitochondrial protein Abcb10 results in altered arginine metabolism in MEL and K562 cells and nutrient stress signaling through ATF4. J Biol Chem. 2023 Jul;299(7):104877. doi: 10.1016/j.jbc.2023.104877. Epub 2023 Jun 1. PMID: 37269954; PMCID: PMC10316008.
Citing Our Facility
Acknowledgments
We would like to thank you for acknowledging the our facility. This recognition allows us to highlight the impact of your work and demonstrates the important contributions of our facility makes to research across the University of Utah. The recognition our core receives from your acknowledgments also aids in receiving grants and further funding for equipment and services we can provide to our users.
Self-Run Services / Instrumentation Usage:
In published papers that used instruments at our facility and notably involved staff members please use the following format:
We acknowledge (facility name) at the University of Utah for use of equipment (insert instrument/service details here), and thank (insert any notable staff member – if desired) for their assistance.
Assisted Services:
In published papers where a staff member assisted you in addition to the requested services please use the following format:
We acknowledge (facility name) at the University of Utah for use of equipment (insert instrument/service details here), and thank (insert staff member-required) for their assistance in (service provided).
Collaboration:
For publications resulting from collaborations that assisted with the methodologies, planning process and execution of your experiment in addition to equipment usage we require Co-author attribution on your publication for our facility and any staff members who provided substantial contributions to the originating project.

