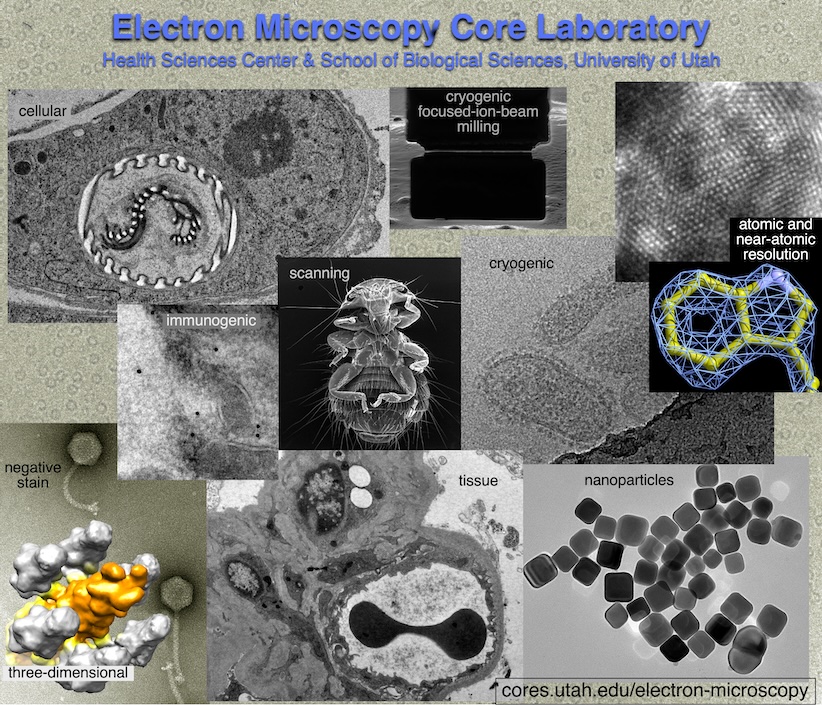
Electron Microscopy
Electron Microscopy prepares and images specimens by transmission or scanning EM and can also do two- and three-dimensional image processing. The lab specializes in biological EM, but also works with non-biological samples. The EM Core Lab can train users in sample preparation, microscope use, and image analysis. Common EM procedures done in the lab include thin-sectioning, negative staining, and cryogenic EM (cryo-EM).
Available Services
EM Imaging
- transmission electron microscopy
- scanning electron microscopy
- room-temperature and cryogenic imaging
- two- and three-dimensional image processing
EM Imaging Instruments
- transmission electron microscopy: JEOL JEM-1400, ThermoFisher Tecnai 20, ThermoFisher Titan Krios
- scanning electron microscopy: Zeiss GeminiSEM-300, ThermoFisher Aquilos 2
Specimen Preparation (EM and light microscopy)
- embedding (in resin)
- thick-sectioning (EM or LM)
- thin-sectioning (EM or LM)
- section staining (EM or LM)
- negative staining (EM)
- sputter coating (EM)
- cryogenic (cryo-EM)
- nanoparticles (EM)
- Tokuyasu (immuno-EM)
- high-pressure freezing (EM)
- freeze substitution (EM)
EM Specimen Preparation Equipment
- ThermoFisher Aquilos 2 (cryogenic focused-ion beam milling)
- ThermoFisher Vitrobot (cryogenic preparation)
- Leica UC7 (cryogenic thick- and thin-sectioning)
- Leica UC6 (thick- and thin-sectioning)
- Leica UCT (thick- and thin-sectioning)
- Rotary Microtome (thick-sectioning of paraffin and plastic)
- Pelco Biowave Microwave Oven (speeds up processing)
- Plasma Glow-Discharger (makes grids hydrophilic)
Training
Training in all equipment, instruments, and techniques mentioned above
Service Rates
Personnel (Staff) Costs
| University of Utah | Outside Academic Institutions | Commercial |
| $50–$85 | $77–$131 | $77–$132 |
Example Prices
Because the time and effort a project takes is variable, we bill according to the time taken and supplies needed. We do not have set fees per sample. Some projects take less time and some take more time than predicted. Typically, preparation of multiple specimens (done simultaneously) will cost less per sample than doing one at a time. Nevertheless, the following are estimated times, supplies, and prices for a few of the analyses we can do in the EM Core Lab. Please contact the lab for direct quotes for your project. Also, please note that we can do only part of what is listed. For example, we could do only imaging or only specimen preparation, and if you prepare a specimen, we can image it, or vice versa.
Thin-Sectioning (one sample)
| Procedure | Components | Univ. of Utah | Outside Acad. Inst. | Commercial |
| Processing | supplies + 3 hr staff time | $194 | $299 | $299 |
| Cutting | 1.5 hr equipment + staff | $150 | $231 | $231 |
| Staining | supplies + 0.5 hr staff | $30 | $47 | $47 |
| Imaging | 2 hr microscope + staff | $200 | $308 | $308 |
| Total | $574 | $885 | $885 |
Negative Staining (one sample)
| Procedure | Components | Univ. of Utah | Outside Acad. Inst. | Commercial |
| Preparation | supplies + 0.5 hr staff time | $46 | $71 | $71 |
| Imaging | 1 hr microscope + staff | $135 | $208 | $209 |
| Total | $181 | $279 | $280 |
Cryogenic Transmission EM (one sample)
| Procedure | Components | Univ. of Utah | Outside Acad. Inst. | Commercial |
| Negative Staining¶ | as above | $181 | $279 | $280 |
| Prepare cryo-specimen | supplies + 1 hr staff | $118 | $182 | $183 |
| Imaging, JEOL 1400 | 1.5 hr microscope + staff | $203 | $312 | $322 |
| Imaging, Krios‡ | 2 hr microsc.,staff,supplies | $283 | $436 | $477 |
| Total, JEOL 1400 | $502 | $773 | $785 | |
| Total, Krios | $582 | $897 | $940 |
¶A check on the sample before the more lengthy and laborious steps of cryogenic EM.
‡Typically, the Krios is used for 24-hour periods, and the per-hour charges can be found on this page. However, we also make occasions for partial-day or hourly use, particularly for screening or short projects.
Scanning EM (one sample)
| Procedure | Components | Univ. of Utah | Outside Acad. Inst. | Commercial |
| Specimen Preparation | supplies + 2 hr staff time | $175 | $270 | $270 |
| Pd-Au coating | supplies | 25 | $39 | $50 |
| Imaging | 1 hr microscope + staff | $90 | $139 | $139 |
| Total | $290 | $448 | $459 |
Pre-Embedding Immunogold EM (one sample)
| Procedure | Components | Univ. of Utah | Outside Acad. Inst. | Commercial |
| Sample Handling† | 3 hr staff time | $150 | $231 | $231 |
| Pre-embedding immunostaining | 3 hr staff time | $150 | $231 | $231 |
| Post-fixation, Resin Embedding | 3 hr staff + equip. | $210 | $324 | $324 |
| Ultramicrotomy, staining | 2 hr staff + equip. | $160 | $247 | $247 |
| Specimen preparation | supplies | $250 | $385 | $385 |
| Imaging | 4 hr staff + equip. | $400 | $616 | $616 |
| Total | $1320 | $2034 | $2034 |
†This is to improve accessibility of antibodies and gold particles.
Tokuyasu Cryo-Sectioning Immunogold EM (one sample)
| Procedure | Components | Univ. of Utah | Outside Acad. Inst. | Commercial |
| Sample Handling | 6 hr staff time | $300 | $462 | $462 |
| Ultramicrotomy, immunostaining | 6 hr staff + equip. | $440 | $678 | $678 |
| Specimen preparation | supplies | $200 | $308 | $308 |
| Imaging | 4 hr staff + equip. | $400 | $616 | $616 |
| Total | $1340 | $2064 | $2064 |
Two-Dimensional Image Processing, Three-Dimensional Image Processing, Data Interpretation, and Report Writing
These would be additional costs charged by the hour for staff time. If you are interested in us doing any of this for you, please inquire about these costs.
Requesting Services
Existing users may login directly to the Resource Scheduling System to schedule or order services. This system is cores-wide and uses University of Utah uNID authentication.
Contact Us
Hours of Operation
9:00 am to 5:00 pm
Monday - Friday
Shipping Address
Electron Microscopy Core Laboratory
2500 Sorenson Molecular Biology Building (SMBB)
36 South Wasatch Drive, Room 2500
Salt Lake City, UT, 84112
Recent Mentions
- Adriaenssens, E., B. Asselbergh, P. Rivera-Mejias, S. Bervoets, L. Vendredy, V. De Winter, K. Spaas, R. de Rycke, G. van lsterdael, F. lmpens, T. Langer and V. Timmerman (2023). Small heat shock proteins operate as molecular chaperones in the mitochondrial intermembrane space. Nat Cell Biol 25(3): 467- 480.10.1038/s41556-022-01074-9
- Ashkarran, A. A., H. Gharibi, J. W. Grunberger, A. A. Saei, N. Khurana, R. Mohammadpour, H. Ghandehari and M. Mahmoudi (2023). Sex-Specific Silica Nanoparticle Protein Corona Compositions Exposed to Male and Female BALB/c Mice Plasmas. ACS Bio Med Chem Au 1(1): 62- 73.10.1021/acsbiomedchemau.2c00040
- Backman, T., S. M. Latorre, L. Eads, S. Som, D. Belnap, A. M. Manuel, H. A. Burbano and T. L. Karasov (2023). A weaponized phage suppresses competitors in historical and modern metapopulations of pathogenic bacteria. bioRxiv: 2023.2004.2017.536465.10.1101/2023.04.17.536465
- Barnard, D. L., D. M. Belnap, P. Azadi, C. Heiss, D. S. Snyder, S. C. Bock and T. W. Konowalchuk (2022). Examining the interactions of Galahad compound with viruses to develop a novel inactivated influenza A virus vaccine. Heliyon 8(7): e09887.10.1016/j.heliyon.2022.e09887
- Beenken, A., G. Cerutti, J. Brasch, Y. Guo, Z. Sheng, H. Erdjument-Bromage, Z. Aziz, S. Y. Robbins-Juarez, E. Y. Chavez, G. Ahlsen, P. S. Katsamba, T. A. Neubert, A. W. P. Fitzpatrick, J. Barasch and L. Shapiro (2023). Structures of LRP2 reveal a molecular machine for endocytosis. Cell 186(4): 821- 836.e813.https://doi.org/10.1016/j.cell.2023.01.016
- Cooney, I., D. C. Mack, A. J. Ferrell, M. G. Stewart, S. Wang, H. M. Donelick, D. Tamayo-Jaramillo, D. L. Greer, D. Zhu, W. Li and P. S. Shen (2023). Lysate-to-grid: Rapid Isolation of Native Complexes from Budding Yeast for Cryo-EM Imaging. Bio Protoc 13(2).10.21769/BioProtoc.4596
- Cooney, I., H. L. Schubert, K. Cedeno, H.-J. L. Lin, J. C. Price, C. P. Hill and P. S. Shen (2023). Visualization of the Cdc48 AAA+ ATPase protein unfolding pathway. bioRxiv: 2023.2005.2013.540638.10.1101/2023.05.13.540638
- Gilcrease, E. B., S. R. Casjens, A. Bhattacharjee and R. Goel (2023). A Klebsiella pneumoniae NDM-1+ bacteriophage: Adaptive polyvalence and disruption of heterogenous biofilms. Front Microbial 14: 1100607.10.3389/fmicb.2023.1100607
- Green, Y. S., M. C. Ferreira Dos Santos, D. G. Fuja, E. C. Reichert, A. R. Campos, S. J. Cowman, K. Acuna Pilarte, J. Kohan, S. R. Tripp, E. A. Leibold, D. Sirohi, N. Agarwal, X. Liu and M. Y. Koh (2022). ISCA2 inhibition decreases HIF and induces ferroptosis in clear cell renal carcinoma. Oncogene 41(42): 4709- 4723.10.1038/s41388-022-02460-1
- Grunberger, J. W. and H. Ghandehari (2023). Layer-by-Layer Hollow Mesoporous Silica Nanoparticles with Tunable Degradation Profile. Pharmaceutics 15(3).10.3390/pharmaceutics15030832
- Momont, C., H. V. Dang, F. Zatta, K. Hauser, C. Wang, J. di lulio, A. Minola, N. Czudnochowski, A. De Marco, K. Branch, D. Donermeyer, S. Vyas, A. Chen, E. Ferri, B. Guarino, A. E. Powell, R. Spreafico, S. S. Yim, D. R. Salce, I. Bartha, M. Meury, T. I. Croll, D. M. Belnap, M. A. Schmid, W. T. Schaiff, J. L. Miller, E. Cameroni, A. Telenti, H. W. Virgin, L. E. Rosen, L. A. Purcell, A. Lanzavecchia, G. Snell, D. Corti and M. S. Pizzuto (2023). A pan-influenza antibody inhibiting neuraminidase via receptor mimicry. Nature 618(7965): 590-597.10.1038/s41586-023-06136-y
- Patel, N., M. A. Johnson, N. Vapniarsky, M. W. Van Bracklin, T. K. Williams, S. T. Youngquist, R. Ford, N. Ewer, L. P. Neff and G. L. Hoareau (2023). Elamipretide mitigates ischemia-reperfusion injury in a swine model of hemorrhagic shock. Sci Rep 13(1 ): 4496.10.1038/s41598-023-31374-5
Citing Our Facility
Acknowledgments
We would like to thank you for acknowledging the our facility. This recognition allows us to highlight the impact of your work and demonstrates the important contributions of our facility makes to research across the University of Utah. The recognition our core receives from your acknowledgments also aids in receiving grants and further funding for equipment and services we can provide to our users.
Self-Run Services / Instrumentation Usage:
In published papers that used instruments at our facility and notably involved staff members please use the following format:
We acknowledge (facility name) at the University of Utah for use of equipment (insert instrument/service details here), and thank (insert any notable staff member – if desired) for their assistance.
Assisted Services:
In published papers where a staff member assisted you in addition to the requested services please use the following format:
We acknowledge (facility name) at the University of Utah for use of equipment (insert instrument/service details here), and thank (insert staff member-required) for their assistance in (service provided).
Collaboration:
For publications resulting from collaborations that assisted with the methodologies, planning process and execution of your experiment in addition to equipment usage we require Co-author attribution on your publication for our facility and any staff members who provided substantial contributions to the originating project.



